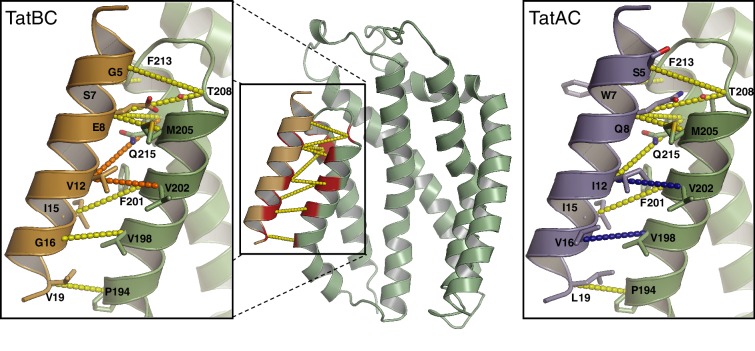
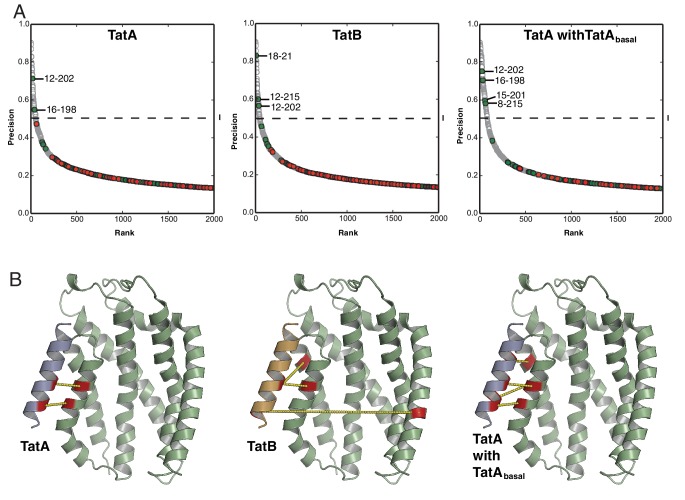
Figure 3. Modeling the interaction of TatA and TatB with the C-terminal end of TatC.
The interaction between the TatA/B TMH and TatC TM5/TM6 modeled for E. coli TatBC and TatAC pairs. The models are based on A. aeolicus TatC crystal packing contacts. Evolutionary couplings for the TatAAll dataset are shown (dotted lines). Couplings retained in just the TatB or TatA datasets with precisions greater than 0.5 (Figure 3—figure supplement 1A) are colored orange or blue, respectively. See also (Table 1).