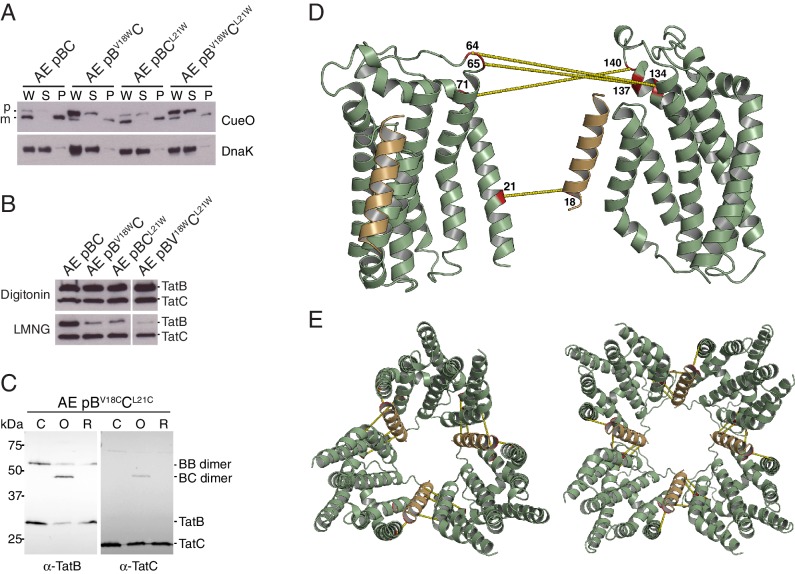
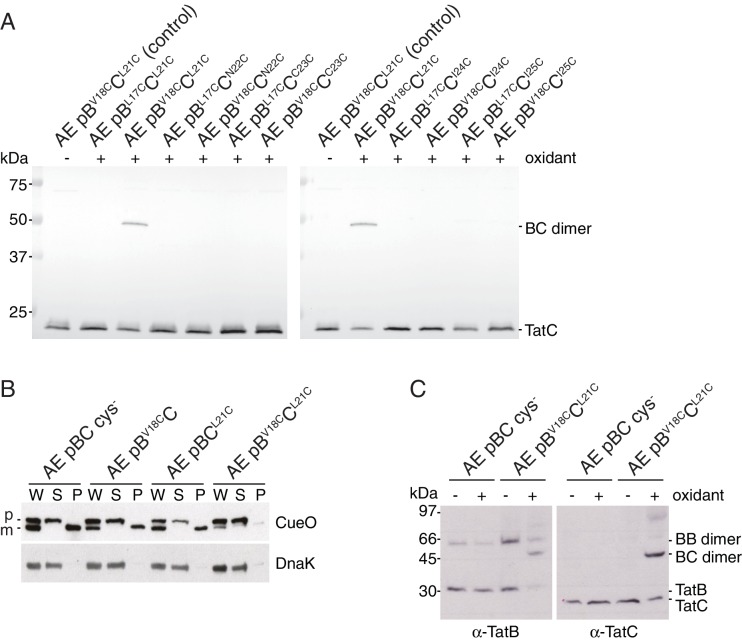
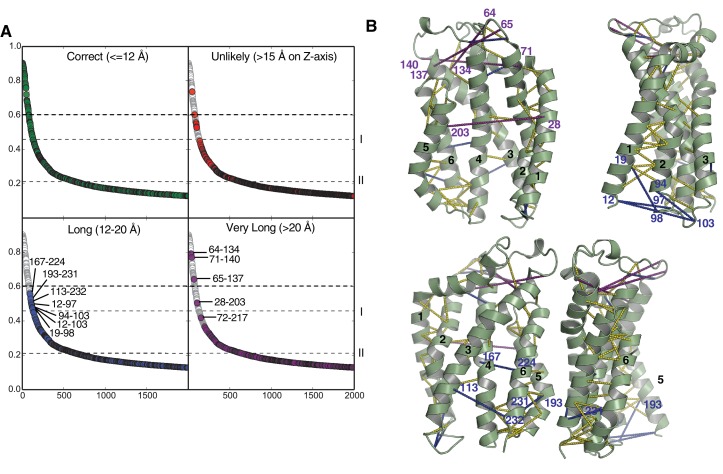
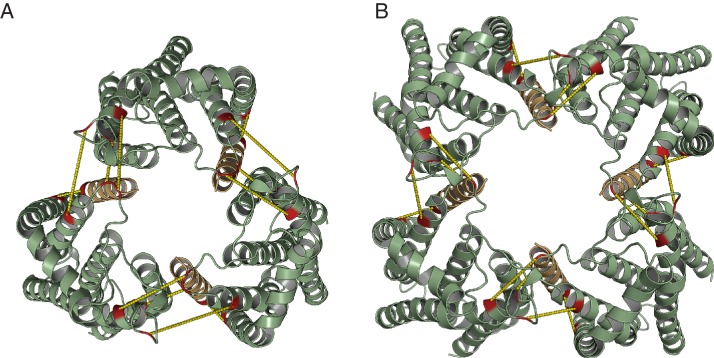
Figure 8. Identification of a second TatBTMH-TatC contact site.
(A) Tat transport activity of strains with tryptophan substitutions targeting the predicted interface between the TatB TMH and TatC TM1. Methodology and labels are as for Figure 5A. (B) Effects of the tryptophan substitutions on TatBC interactions. Cell lysates were solubilized in either digitonin (top panel) or LMNG (bottom panel), immunoprecipitated with antibodies against TatC, and then immunoblotted with a combination of TatB and TatC antibodies. (C) Disulfide crosslinks can be detected at the predicted interface between the TatB TMH and TatC TM1. Cells carrying the indicated cysteine substituted Tat variants were subjected to a mock incubation (‘C’, no oxidant or reductant), oxidizing (‘O’, copper phenanthroline) or reducing (‘R’, DTT) conditions. Membranes were then isolated and subjected to immunoblotting with TatB (left panels) or TatC (right panels) antibodies. (D) Structural representation of the highest-scoring co-evolution-predicted contacts between TatBTMHC heterodimers (precision >0.6). (E) Model for the TatBC complex based on docking either three (Left) of four (Right) TatBTMH-TatC heterodimers to optimize agreement with the co-evolution data in (D). The complexes are viewed from the cytoplasmic side of the membrane. See also Figure 8—figure supplement 3, Video 3 and Supplementary files 1 and 2.