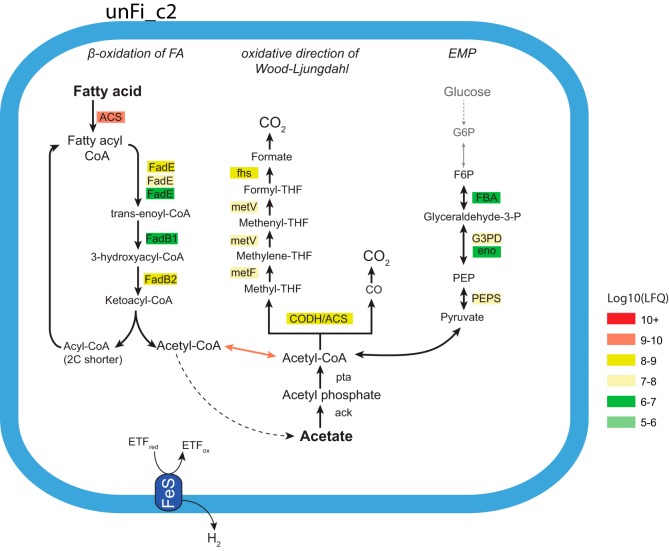
FIG 3.
Selected metabolic pathways of the putative novel SAOB unFirm02_FrBGR cluster 2. The pathways are proposed based on genome and proteome comparison, and protein abundances are indicated by color, ranging from high abundance (red) to low abundance (green). All enzymes needed for β-oxidation of fatty acids, in addition to most enzymes associated with the Wood-Ljungdahl pathway, were detected for unFirm02_FrBGR cluster 2 (unFi_c2). Parts of both pathways were also detected for unFirm02_FrBGR cluster 1 (unFi_c1), albeit at a lower detection level (see Fig. S4 in the supplemental material). Only proteins affiliated to the lower part of EMP were represented in the proteome. Acetate kinase (ack) was detected in the genome but not in the proteome. More details on the proteins detected can be found in Table S3, including abbreviations.