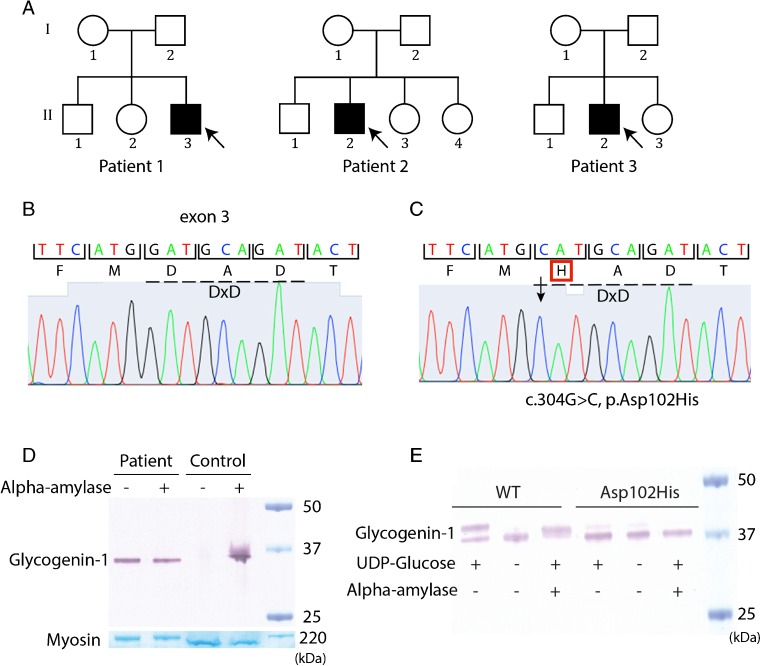
Fig. 2.
Genetic and protein analysis. a Pedigrees of the families. b Chromatogram of normal GYG1 DNA sequence with the DxD motif indicated by hatched line. c Chromatogram demonstrating a homozygous missense mutation in GYG1 (c.304G > C, p.Asp102His), the hatched line shows that the DxD motif is mutated. d Western blot analysis of glycogenin-1 on protein extracted from cardiac muscle biopsy specimen from patient 1 and a control sample performed without (−) or with alpha-amylase (+) treatment. In patient 1 glycogenin-1 was detected both without and with alpha-amylase treatment compared to control sample where glycogenin-1 was only detected after alpha-amylase treatment. The band corresponding to myosin heavy chain was used as loading control. e Cell-free autoglycosylation in vitro revealed that the protein with the mutation p.Asp102His was not able to autoglycosylate demonstrated with only one band after addition of uridine diphoshate (UDP) glucose (+) compared to wild type (WT) protein which was able to autoglycosylate demonstrated with two bands corresponding to both unglucosylated (lower band) and glycosylated (upper band) glycogenin-1. Unglucosylated glycogenin-1 weighs approximately 1 kDa less than free autoglucosylated protein. After alpha-amylase treatment the short glucose polymers are cleaved off resulting in only one band for WT compared to p.Asp102His which was not affected because of absence formation of short glucose polymers