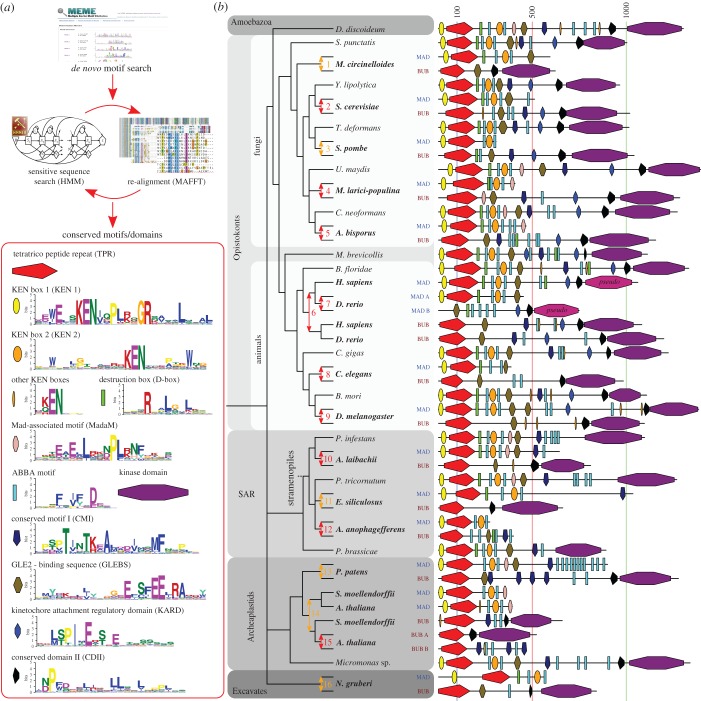
Figure 1.
Fate of conserved functional sequence features after 16 independent duplications of the MADBUB gene family throughout eukaryotic evolution (a) Overview of the de novo sequence discovery pipeline ConFeaX including the ancestral conserved features of a search against the eukaryotic MADBUB gene family. The consensus sequences of the detected conserved motifs are depicted as a sequence logo (colours reflect distinct amino acid properties and height of the letters indicates conservation of amino acids). Each feature is assigned a differently coloured shape. (b) Cartoon of the evolutionary scenario of 16 independent duplications of the MADBUB gene family throughout eukaryotic evolution, including a projection of conserved features onto the linear protein representation (on scale). Gene duplications are indicated by an arrow (red: high confidence, orange: likely). The subfunctionalized paralogues MAD and BUB are coloured brown and blue, respectively. Numbers indicate the clades in which the duplications occurred: 1, Mucorales; 2, Saccharomycetaceae; 3, schizosaccharomycetes; 4, pucciniomycetes: 5, agaricomycetes (excluding early branching species); 6, vertebrates; 7, teleost fish; 8, nematodes; 9, diptera (flies); 10, Albuginaceae (oomycete); 11, Ectocarpales (brown algae); 12, Aureococcus (harmful algae bloom); 13, bryophytes (mosses); 14, tracheophytes (vascular plants); 15, magnoliaphytes (flowering plants); 16, Naegleria.