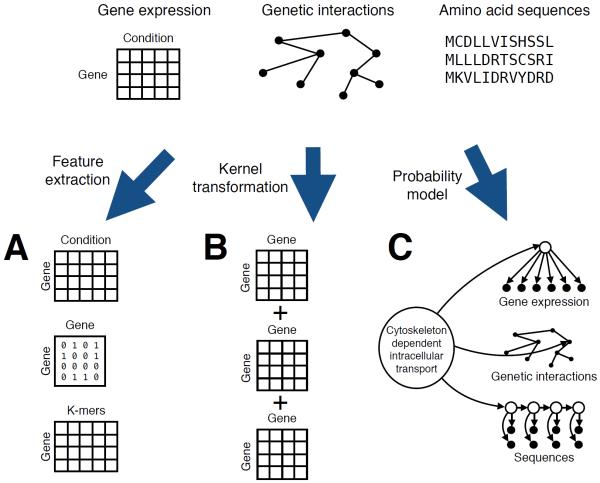
Figure 5. Three ways to accommodate heterogeneous data in machine learning.
The task of predicting gene function labels requires methods that take as input gene expression data, protein sequences, protein-protein interaction networks, etc. These diverse data types can be encoded into fixed-length features, represented using pairwise similarities (kernels), or directly accommodated by a probability model.