Abstract
Prions are proteins that can adopt self-perpetuating conformations and are traditionally regarded as etiological agents of infectious neurodegenerative diseases in humans, such as Creutzfeldt–Jakob disease, kuru, and transmissible encephalopathies. More recently, a growing consensus has emerged that prion-like, self-templating mechanisms also underlie a variety of neurodegenerative disorders, including amyotrophic lateral sclerosis, Alzheimer’s disease, and Huntington’s disease. Perhaps most surprising, not all prion-like aggregates are associated with pathological changes. There are now several examples of prion-like proteins in mammals that serve positive biological functions in their aggregated state. In this review, we discuss functional prions in the nervous system, with particular emphasis on the cytoplasmic polyadenylation element-binding protein (CPEB) and the role of its prion-like aggregates in synaptic plasticity and memory. We also mention a more recent example of a functional prion-like protein in the brain, TIA-1, and its role during stress. These studies of functional prion-like proteins have provided a number of generalizable insights on how prion-based protein switches may operate to serve physiological functions in higher eukaryotes.
Prions and prion-like mechanisms are typically regarded as disease-causing agents in animals. However, there are now examples of prion-like proteins that serve positive functions (e.g., CPEB in synaptic plasticity and memory).
In mammals, prions are classically defined as protein-based agents of infectious neurodegenerative disease (Prusiner 1982; Weissmann 2004). However, there is now evidence that a variety of neurodegenerative conditions that are not transmissible between individuals may nevertheless be caused by self-templating, prion-like mechanisms (Prusiner 2013). The pathogenic proteins associated with these disorders—for example, amyloid β in Alzheimer’s disease and huntingtin in Huntington’s disease—undergo spontaneous conversion from a soluble conformation to a self-propagating, aggregated structure that is cytotoxic. In contrast, yeast prions are characterized by their ability to propagate phenotypes from mother to daughter cell in a stable, non-Mendelian manner and also without involvement of nucleic acid–based mechanisms.
Although the distinction between these definitions is often semantic and increasingly blurred, evidence from a number of recent studies now supports the idea that “prion-like” proteins are more ubiquitous than originally thought and may perform a wide range of biologically meaningful roles in both simple and higher eukaryotes. Furthermore, there is growing evidence that prion-like proteins can serve positive functions in their aggregated state. We use the term “prion-like” to indicate proteins that share several properties with yeast prions as well as disease-causing prions in animals, including detergent resistance, protease resistance, self-propagation, and transmissibility. We use the term “functional” to describe the observation that these prion-like proteins serve normal physiological functions in their aggregated state, a concept that has gained traction in a variety of different biological contexts.
A compelling example of a functional prion-like protein in mammals is the immunological protein MAVS. In 2011, Hou et al. (2011) found that MAVS, a mitochondrial protein that activates the transcription factors IRF3 and NF-κB during viral infection and thus induces type 1 interferons, forms prion-like aggregates that activate and propagate antiviral innate immune responses. In its aggregated state, MAVS activates IRF3, whereas MAVS fibrils formed in vitro efficiently promote a prion-like mechanism that causes the aggregation of the endogenous protein, although it remains to be determined whether cell-to-cell transmission can occur (Hou et al. 2011). It is conceivable that prion-like aggregation of MAVS facilitates a robust and highly sensitive response to viral infection, but otherwise imposes a formidable kinetic barrier that guards against spurious, potentially detrimental activation of innate immunity in the absence of viral infection. Moreover, prion-like aggregation may give rise to complex structural determinants that are not easily exploited by viral mechanisms designed to circumvent host immunity. Thus, MAVS shows prion-like properties but, unlike conventional prions, serves a positive function in the aggregated state, which is not associated with disease.
In this article, we focus on functional prion-like proteins in the central nervous system and, in particular, on the cytoplasmic polyadenylation element-binding protein (CPEB). We examine how conversion from a soluble to a prion-like conformation regulates protein synthesis at the synapse, thereby modulating both the maintenance of synaptic plasticity and long-lasting memory. The small but growing list of functional prion-like proteins in animals gives credence to the idea that prion-like aggregation serves a physiological role in a variety of disparate biological contexts and, importantly, is regulated by the cell, in stark contrast to the behavior of conventional prions, whose conversion is spontaneous and unregulated. Indeed, the ubiquity of amyloid structures found in both prions and nonprions alike, along with the capacity of many proteins to form prion-like aggregates, suggests that prion diseases may actually represent a rare consequence of a common structural motif that has been co-opted in nature for pathogenic functions.
THE MAINTENANCE OF LONG-TERM MEMORY
To investigate the role of CPEB and its prion-like properties in the context of long-term memory storage, we first describe the neural circuits in the marine sea snail Aplysia californica used to study learning and memory. We then describe the experiments that led to the identification and characterization of CPEB in the brain.
Aplysia has a simple nervous system that makes it highly amenable for studying the neuronal mechanisms of reflexes at the cellular and molecular levels (Kandel 2001). Specifically, Aplysia has a number of simple reflexes that can be modified by different forms of learning. A classic example is the gill-withdrawal reflex. The gill is an external respiratory organ that is covered by a sheet of skin called the mantle shelf, which terminates in a fleshy spout called the siphon. A weak tactile stimulus applied to the siphon elicits a brisk withdrawal of the gill for protection underneath the mantle shelf. This withdrawal can be modified by various forms of learning, including sensitization, which is a form of learned fear. A shock applied to the tail scares the animal, such that the same weak tactile stimulus that had produced a modest withdrawal of the gill in the resting snail now produces a much more vigorous, longer-lasting withdrawal. Moreover, the duration of the memory for this noxious event varies as a function of a number of training trials. Thus, a single tail shock produces a short-term form of sensitization that lasts for minutes, but a series of five shocks produces a long-term form of sensitization that lasts for days or weeks.
Work on this reflex first showed that synaptic plasticity, the ability of neurons to modulate the strength and structure of their synapses, is a key mechanism contributing to learning and memory storage. The neural circuit of the gill-withdrawal reflex has an important monosynaptic component in which the sensory neurons that innervate the siphon make direct connections to the motor neurons that move the gill. Like behavioral memory, synaptic plasticity between the sensory and motor neurons has at least two temporally distinct forms: a short-term form lasting minutes and a long-term form lasting days or weeks. These temporally distinct forms of learning-related synaptic plasticity have distinct molecular requirements. The short-term form depends on the covalent modifications of preexisting proteins mediated in part by protein kinase A (PKA) that lead to the strengthening of preexisting connections. In contrast, the long-term form requires both new protein synthesis and CREB-mediated gene expression, which drive the remodeling of preexisting synapses and the formation of new synaptic connections.
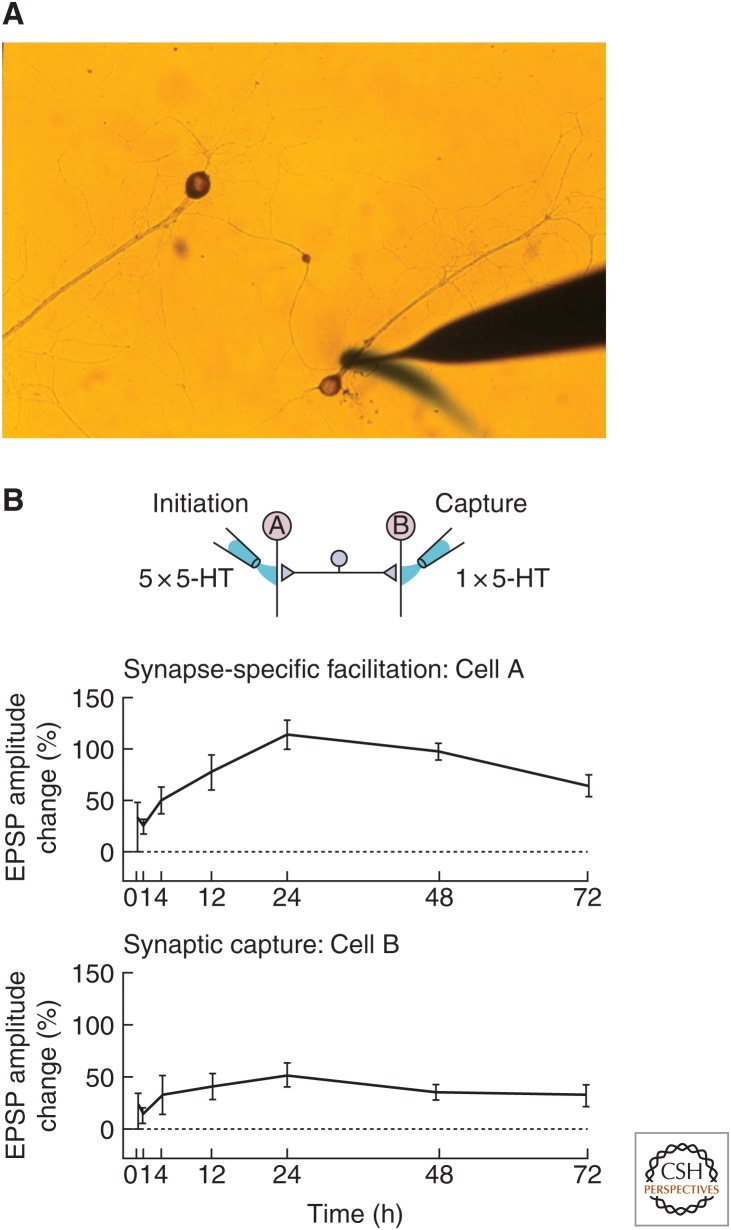
Because long-term synaptic plasticity requires transcription and therefore the nucleus, which, in principle, is accessible by every synapse of the neuron, it raises the question of whether all long-term changes must necessarily be cell-wide or whether long-term changes can be transcriptionally dependent, yet restricted to some synapses and not to others. To address this problem, Martin et al. (1997) developed a new culture system in Aplysia in which a single bifurcated sensory neuron of the gill-withdrawal reflex was plated in contact with two spatially separated gill motor neurons (Fig. 1A). In this culture system, application of a single pulse of 5-HT (serotonin, a neuromodulatory transmitter released in vivo by interneurons activated during learning) to one of the two sets of synapses results in a synapse-specific short-term facilitation of preexisting connections that lasts for minutes. Application of five pulses of 5-HT to one of the two sets of synapses, which simulates the spaced behavioral training that leads to long-term memory, also elicits branch-specific long-term facilitation that lasts for 3 or more days (Fig. 1B). Although the short-term form does not require new protein synthesis, the long-term form requires both transcription in the nucleus and local protein synthesis at activated synapses. Moreover, the long-term facilitation initiated in one branch of the bifurcated preparation can be captured at the other branch by applying only a single pulse of 5-HT (which by itself is capable of producing only branch-specific short-term facilitation). This finding first suggested that gene products are sent to all the synapses of the neuron, but only those that are marked by the modulatory transmitter 5-HT can use these gene products productively.
Figure 1.
Long-term facilitation induced by serotonin (5-HT) at one branch of an Aplysia bifurcated sensory-motor neuron coculture can be captured at the other branch. (A) Microcultures of individual neurons of the gill-withdrawal reflex for studying branch-specific plasticity (Casadio et al. 1999). Here, one branch of the bifurcated sensory neuron is perfused with 5-HT. The cell body of the sensory neuron is visible in the center of the image. (B) In synapse-specific facilitation, five spaced pulses of 5-HT to one branch of a bifurcated sensory-to-motor neuron Aplysia coculture leads to long-term facilitation lasting at least 72 h. A single pulse of 5-HT to the other branch, which alone normally produces only short-term facilitation lasting minutes, now recruits long-term plasticity in a process called synaptic capture.
These experiments, as well as parallel experiments in the hippocampus (Frey and Morris 1997), suggest that individual sets of synapses can independently undergo long-term synaptic changes, both structural and electrophysiological, and these changes can persist for more than 24 h. Thus, long-term facilitation and its associated structural changes are specific only to stimulated (and therefore marked) synapses. Moreover, this branch-specificity requires CREB-1 activation in the presynaptic sensory neurons. Thus, signals are sent not only from the synapse back to the nucleus (Martin and Kandel 1996; Lee et al. 2007), but also from the nucleus to specific synapses. In other words, these results indicate that the molecular dialogue required for synapse-specific long-term memory is bidirectional.
What is the molecular nature of the synaptic mark? Casadio et al. (1999) examined this question and found that the mark has at least two components: (1) a PKA-dependent component needed for the initial capture of synapse-specific facilitation and for the growth of new synaptic connections, which is revealed by perfusing a single branch of sensory-to-motor neuron cocultures with a nonhydrolyzable analog of cAMP; and (2) a local protein synthesis-dependent component needed for the maintenance of long-term facilitation and for the stabilization of synaptic growth beyond 24 h, which is revealed by treatment with protein synthesis inhibitors such as emetine. Because mRNAs are made in the cell body and then presumably distributed to all terminals, the need for local translation of some mRNAs only at a specific set of synapses and not at others suggested that these mRNAs may be dormant before they reach the site of translation (Si et al. 2003a). If so, then the synaptic mark for stabilization might be a regulator of translation capable of activating translationally dormant mRNAs.
IDENTIFICATION AND CHARACTERIZATION OF APLYSIA CPEB
In searching for such a translational regulator that could help explain the phenomenon of synaptic tagging and capture, Si and Kandel (2016) focused on CPEB, a protein that Joel Richter found to activate dormant mRNAs in other biological contexts (McGrew et al. 1989; Si et al. 2003a,b). Earlier work on Xenopus oocytes revealed that some translationally dormant mRNAs are activated following elongation of their poly(A) tail (McGrew et al. 1989; Stebbins-Boaz et al. 1996; Schroeder et al. 1999). This polyadenylation-dependent translational control requires a cis-acting cytoplasmic polyadenylation element (CPE) in the 3′ UTR of the mRNA to which CPEB can bind and recruit a variety of partner proteins (Fox et al. 1989; Hake and Richter 1994; Gebauer and Richter 1996).
In addition to its ability to activate dormant mRNAs, CPEB has three additional features that made it an attractive candidate as a synapse-specific mark for stabilizing synaptic plasticity: (1) it is activated through an extracellular signal, (2) its localization can be spatially restricted (Bally-Cuif et al. 1998; Schroeder et al. 1999; Tan et al. 2001), and (3) a number of mRNAs targeted by CPEB are involved in the cellular growth required for the formation of new synaptic connections during long-term memory storage growth (Chang et al. 2001; Groisman et al. 2002).
Si et al. (2003a,b) proceeded to identify two homologs of CPEB in Aplysia. One was the developmental form of CPEB that Hake and Richter (1994) had studied, but they also found a new isoform that they named ApCPEB. Si et al. (2003a) found that ApCPEB can be activated at the synapse by a single pulse of 5-HT. Moreover, blocking this form of CPEB by means of an antisense oligonucleotide at a marked (active) synapse simulated the action of protein synthesis inhibitors applied to the active synapse. It did not affect the initiation of long-term synaptic facilitation, but it blocked the maintenance of long-term facilitation a day or so after the long-term memory was formed (Si et al. 2003a).
A striking feature of ApCPEB is that its N-terminus is highly enriched in the polar residues glutamine and asparagine, which is a characteristic of the prion domain of yeast prions. This observation immediately raises the intriguing possibility that ApCPEB may use prion-like properties to facilitate stable patterns of gene expression in a synapse-specific and activity-dependent manner. This made it tempting to think that a prion-like mechanism could underlie synaptic tagging and capture.
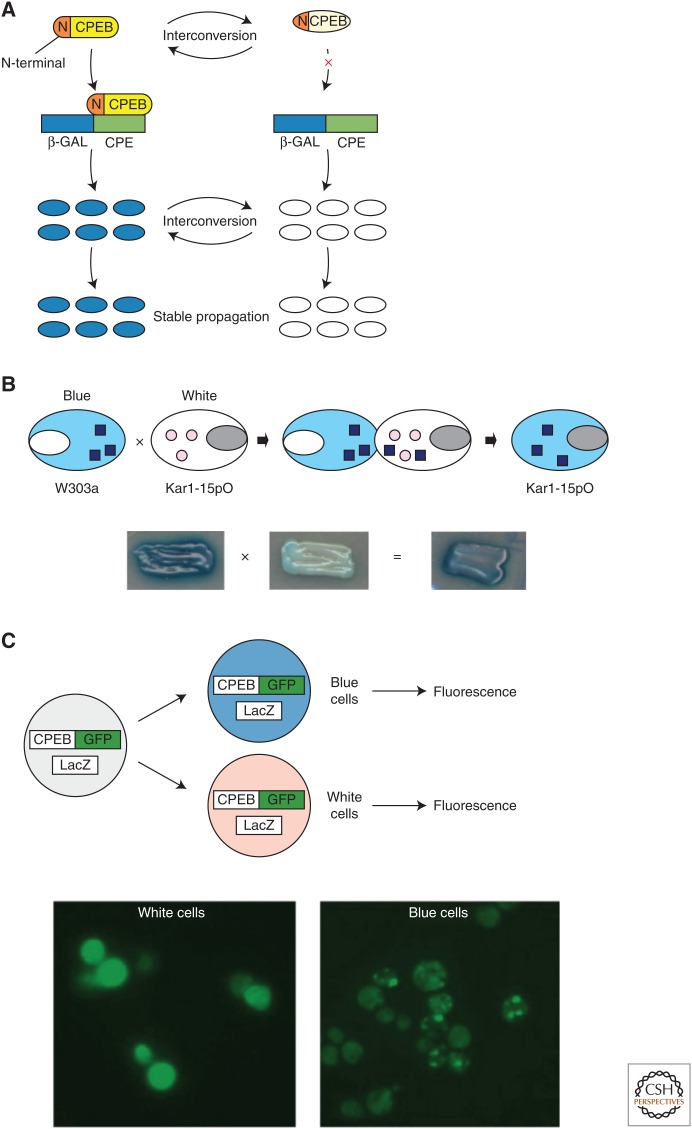
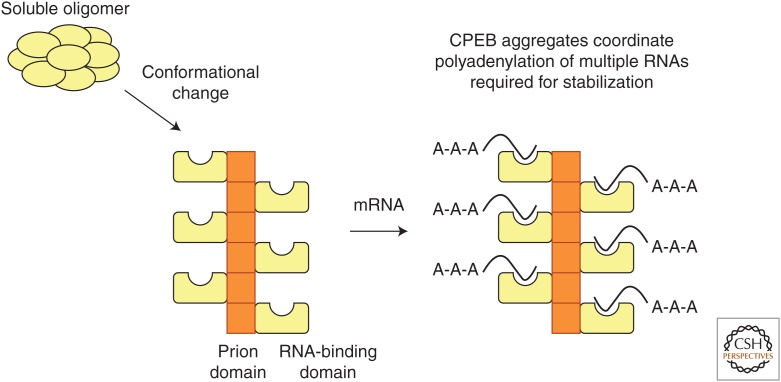
As Prusiner (1982) first showed, prion-like proteins are self-replicating structures. Indeed, Si et al. (2003b) found in both yeast and Aplysia that the N-terminal prion domain endows ApCPEB with self-sustaining properties, and the full-length protein can exist in two functional conformational states: monomeric and aggregated. In a heterologous assay in yeast, the aggregated form of ApCPEB proved to be heritable (Fig. 2A) and could recruit and convert monomers to the aggregated state in cytoduction assays (Fig. 2B). However, unlike prions found in higher eukaryotes, CPEB in its aggregated conformation does not kill cells. In fact, it is the aggregated conformation that is the active form of the protein that promotes translation of target mRNAs (Fig. 2C). Taken together, these observations are consistent with the idea that ApCPEB is a functional prion-like protein, thus prompting the question of how ApCPEB functions in neurons, whether it undergoes neuron-to-neuron transmissibility, and whether its prion-like properties can be used to establish molecular memory.
Figure 2.
Aplysia cytoplasmic polyadenylation element-binding protein (ApCPEB) has properties of a prion in yeast. (A) The full-length CPEB can exist in two functional conformational states that are both interconvertible and heritable. The aggregates enable dominant cytoplasmic inheritance of the blue state. (B) Protein from blue cells recruits and converts the protein from the white cells in a self-perpetuating manner. (C) Aplysia CPEB-GFP (green fluorescent protein) forms aggregates in some cells, but not in others (green signal). However, only aggregated CPEB-GFP is associated with translation of a cotransfected LacZ construct under control of a CPEB response element (blue cells). That is, only the aggregated form of the protein is translationally active.
TOWARD A MODEL FOR NEURONAL CPEB FUNCTION
Si et al. (2010) proposed that, in a naïve synapse, ApCPEB is monomeric and either less active or acts as a repressor, whereas synaptic activation leads to the conversion of ApCPEB to a dominant, self-perpetuating, multimeric state. However, unlike conventional prions, the dominant state of ApCPEB is either more active or devoid of the repressive function of the basal state. Moreover, this dominant state is locally restricted and creates a self-sustaining synaptic mark limited to the specific set of synapses that have been activated. Thus, a sustained period of translation ensues at activated synapses, enabling long-term maintenance of functional and structural synaptic changes.
Testing this model requires an experimental analysis in neurons that addresses four questions. First, does ApCPEB have the properties of a self-perpetuating prion-like protein in neurons? Second, is the prion-like conversion from a soluble to an aggregated form modulated by synaptic stimulation? Third, is ApCPEB active in the aggregated, prion-like state in neurons? Finally, is the prion-like state of ApCPEB required for the maintenance of long-term facilitation? In the presynaptic sensory neurons of Aplysia, both the exogenously expressed and the endogenous Aplysia CPEB exist in two states, one of which is multimeric (Si et al. 2010). Fluorescence complementation experiments indicate that exogenously expressed multimers of ApCPEB in the sensory neuron are likely to result from a homotypic interaction (Si et al. 2010). Moreover, live-cell imaging of the composition of CPEB aggregates shows their self-sustaining nature. Specifically, ectopic expression of CPEB tagged with a photoconvertible fluorescent protein reveals that aggregates can recruit newly synthesized protein in a self-perpetuating manner (Si et al. 2010). These features are consistent with ApCPEB having prion-like properties in the sensory neuron. Notably, CPEB aggregation is lost by deletion of the N-terminal prion domain, demonstrating the critical structural importance of this domain (Si et al. 2010).
AGGREGATION OF APLYSIA CPEB IS INDUCED BY 5-HT
Most examples of prion-like conversions are stochastic in nature. Si and Kandel (2016) postulated that the conversion of ApCPEB to the multimeric state should be regulated by synaptic activity. To address this question, Si et al. (2010) expressed enhanced green fluorescent protein (EGFP)-tagged ApCPEB in the sensory-to-motor neuron coculture, waited 48 h for the expression to reach a steady-state level, and then stimulated the cells with five pulses of 5-HT, the neuromodulatory transmitter released during behavioral learning. In stimulated neurons, they observed a dramatic increase in the number of detectable fluorescent puncta compared to the unstimulated cells, indicating that aggregation of ApCPEB is induced by a physiological signal.
INHIBITING THE ACTIVITY OF THE AGGREGATED FORM OF ApCPEB BLOCKS PERSISTENCE OF SYNAPTIC FACILITATION
If ApCPEB is an active protein in its aggregated conformation, then one would predict that blocking the endogenous form of aggregated ApCPEB should destabilize the maintenance of long-term facilitation. To test this prediction, Si et al. (2010) took advantage of an antibody (Ab464) that preferentially binds the multimeric form of Aplysia CPEB, under the assumption that binding of antibody to the protein would either inhibit or diminish its activity. Injection of this antibody into the sensory cell of sensory-to-motor neuron cocultures did not interfere with basal synaptic transmission or with long-term facilitation measured at 24 h post-bath application of five pulses of 5-HT. However, unlike the control neurons, synaptic facilitation failed to persist beyond 48 h in Ab464-injected cells. These results support the notion that the multimeric form of ApCPEB is involved in long-term stabilization of activity-dependent change in synaptic efficacy.
Taken together, these studies suggested that self-perpetuating conformational changes reminiscent of prion conversion to the “infectious scrapie” form might underlie the ability of ApCPEB to perpetuate memory in an epigenetic fashion. Learning-induced activation of ApCPEB initiates a local positive feedback loop within the synapse, whereby ApCPEB promotes the activation of more ApCPEB through specific conformational changes. This is potentially a powerful mechanism for generating a self-perpetuating local signal at the synapse.
Orb2 IS AN ApCPEB HOMOLOG IN DROSOPHILA
With several of these questions in mind, Kausik Si began to explore CPEB in the Drosophila brain. The Drosophila CPEB family is comprised of two genes, Orb1 and Orb2. Orb1 is an ortholog of mammalian CPEB1, whereas Orb2 is more similar to the mammalian CPEB-2 and -3 (Keleman et al. 2007). Orb proteins share common RNA-binding motifs in the C-terminal end but differ significantly in the N-terminal end, suggesting functional differences among isoforms. Similar to ApCPEB, Drosophila Orb2 has a prion-like domain at the N-terminus, whereas the Orb2 prion domain can functionally substitute the canonical prion domains of yeast prion Sup35.
As is true of ApCPEB, Orb2 has two distinct physical states in the adult fly brain: a monomeric state and a stable, amyloid-like oligomeric state resistant to the detergent sodium dodecyl sulfate (SDS). The oligomers are formed at physiological concentrations of Orb2 protein, induced by stimulation of behaviorally relevant neurons such as the Kenyon cells of the mushroom body or octopamine or dopamine responsive neurons, which increase the level of the oligomeric Orb2 (Krüttner et al. 2012; Majumdar et al. 2012). Considering the evolutionary proximity between Aplysia CPEB and Drosophila Orb2, these observations suggest that the amyloidogenic oligomers of Orb2/CPEB may act to stabilize activity-dependent changes in synaptic efficacy across species.
ROLE OF THE PRION-LIKE STATE OF DROSOPHILA Orb2 IN THE PERSISTENCE OF MEMORY
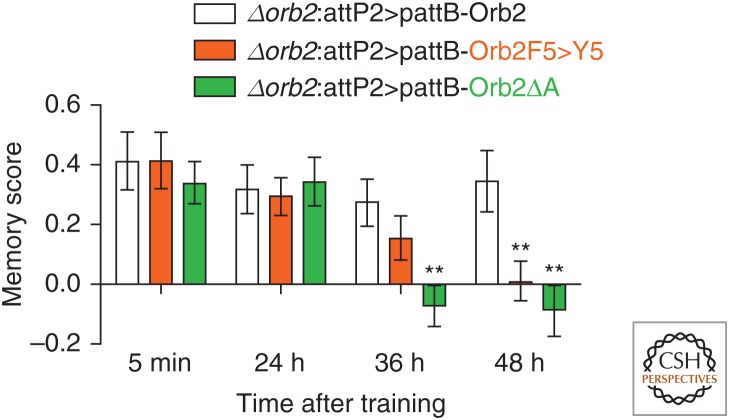
Drosophila Orb2 has two protein isoforms, Orb2A and Orb2B, which differ in their biophysical properties but can form hetero-oligomers. Orb2A is more efficient at forming amyloids than Orb2B, both in vitro and in vivo. Although Orb2A is present at a very low level, its deletion prevents Orb2B oligomerization, suggesting that the rare Orb2A protein either acts directly as a seed to induce activity-dependent oligomerization of the abundant Orb2B protein, or that Orb2A oligomerization indirectly affects oligomerization of Orb2B. A random mutagenesis screen led to the generation of several mutations in Orb2A that prevented its oligomerization (Majumdar et al. 2012). One of these point mutations in Orb2A, Orb2F5 > Y5, reduced activity-dependent amyloid-like oligomerization of Orb2 when introduced into the fly. Notably, flies carrying this mutation showed a very specific memory deficit. Specifically, in two different behavioral paradigms, appetitive associative memory (Fig. 3) and male courtship suppression memory (not shown), the memory scores of mutant flies were similar to those of wild-type flies a day after training. However, unlike wild-type flies, the mutants show a decay in memory at 36 h and by 48 h, it has disappeared altogether. The loss of memory in the Orb2 mutant that cannot form Orb2 amyloid is reminiscent of the loss of facilitation in sensory-to-motor neuron synapses on inhibition of the ApCPEB oligomers via antibody. These data support the notion that prion-like conversion of Orb2 is necessary for long-term memory.
Figure 3.
Orb2 is a prion-like protein in Drosophila whose aggregation is critical for the maintenance of long-term memory. A point mutation in Orb2A (Orb2F5 > Y5) reduces activity-dependent oligomerization of Orb2. Flies harboring this mutation or complete deletion (Orb2ΔA) do not show long-term memory in an appetitive associative memory task.
CPEB IN THE MAMMALIAN BRAIN
More recently, Fioriti et al. (2015) asked whether the function of CPEB in regulating persistent changes in synaptic plasticity in Aplysia is also important in the mammalian nervous system. Thus, they searched for CPEB orthologs in the mouse and identified four isoforms, CPEB-1 to CPEB-4. Of these, CPEB-3 is most similar to ApCPEB (Theis et al. 2003) because it is a neuronal-specific translational regulator with an unstructured Q/N-rich domain at its N-terminus and RNA-binding domains at its C-terminus. These studies in hippocampal neurons provide evidence that mCPEB-3, like its invertebrate orthologs ApCPEB and Orb2, displays hallmark features of prion proteins, including the formation of SDS-resistant aggregates and, in heterologous yeast assays, heritability (Stephan et al. 2015). Furthermore, in the latter study, deletion analysis of CPEB-3 uncovered a novel, tripartite organization in which two aggregation-promoting domains are flanked by a regulatory module that affects interaction between CPEB-3 and actin cytoskeleton. Collectively, these data suggest that CPEB-3 is a functional prion-like protein in the mammalian brain and highlight the potential importance of an actin/CPEB-3 feedback loop for the synaptic plasticity underlying the persistence of long-term memory.
What is the function of CPEB-3 aggregation in the mouse brain? In the basal state, CPEB-3 binds to and represses the translation of target mRNAs such as the AMPA receptor subunits GluA1 and GluA2 (Huang et al. 2006; Pavlopoulos et al. 2011; Fioriti et al. 2015). When CPEB-3 is activated by specific posttranslational modifications during learning, CPEB-3 transitions from a soluble to an aggregated form, which in turn promotes the translation of the AMPA receptor.
THE PERSISTENCE OF SYNAPTIC PLASTICITY AND MEMORY STORAGE REQUIRES CPEB-3-MEDIATED PROTEIN SYNTHESIS IN THE HIPPOCAMPUS
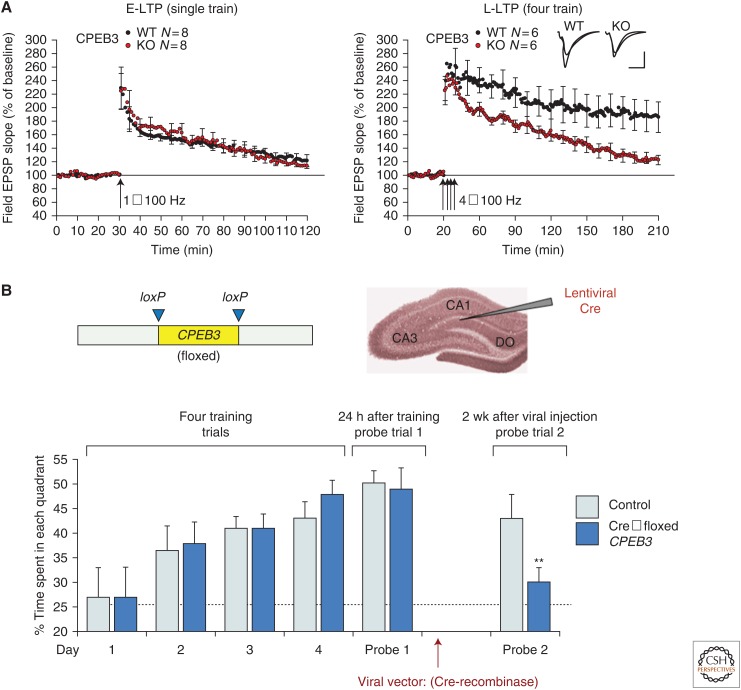
To determine the role of CPEB-3 in the persistence of synaptic plasticity and memory, a conditional knockout (KO) strain of CPEB-3 was generated (Fioriti et al. 2015). These studies revealed that CPEB-3-mediated protein synthesis is required for the maintenance of protein synthesis–dependent long-term potentiation (LTP), but not for short-term LTP or memory acquisition (Fig. 4A). The electrophysiological findings in CPEB-3 conditional KO mice are accompanied by a memory deficit in two different behavioral paradigms—spatial object recognition and the Morris water maze task (Fig. 4B)—suggesting that CPEB-3-mediated processes are required for the maintenance of different types of hippocampal-based spatial learning tasks generated (Fioriti et al. 2015). Furthermore, CPEB-3 loses its ability to maintain long-term synaptic plasticity and long-term memory if its N-terminus, which is necessary for aggregation, is deleted (Fioriti et al. 2015). Thus, like ApCPEB and Drosophila Orb2A, CPEB-3 ostensibly promotes the maintenance of long-term memory through a stimulus-induced conformation change, which in turn leads to protein aggregation. In the case of CPEB-3, this aggregated state is accompanied by a change in function that allows enhanced translation of target mRNAs, such as the AMPA receptor subunits GluA1 and GluA2. These results provide the first evidence for a prion-like mechanism to sustain memory in the mouse brain, during both consolidation and maintenance.
Figure 4.
Mouse CPEB-3 is required for both normal long-term potentiation in the hippocampus and maintenance of long-term spatial memory. (A) Mouse CPEB-3 is required for the maintenance of protein synthesis–dependent long-term potentiation (LTP) induced by strong tetanic stimulation (4 × 100 Hz), but is dispensable for early-phase LTP induced by a single 100 Hz train. (B) Conditional deletion of CPEB-3 by injection of a Cre recombinase lentivirus into the mouse hippocampus prevents reconsolidation following memory retrieval.
AGGREGATION OF FUNCTIONAL PRIONS IS REGULATED
Unlike pathological prions, aggregation of CPEB proteins is a process that is actively regulated. For example, the propensity of CPEB to transition from a soluble to aggregated conformation is intrinsically dependent on protein expression level. As mentioned earlier, transcription of ApCPEB is upregulated in response to 5-HT, leading to elevated protein expression and subsequent aggregation by a variety of structural mechanisms (Raveendra et al. 2013; Fiumara et al. 2015). Moreover, CPEB expression is regulated by microRNAs (miRNAs). In particular, a neuron-specific miRNA, miRNA-22, inhibits ApCPEB mRNA through multiple 3′ UTR binding sites and is downregulated by serotonin (Fiumara et al. 2015). Inhibition of miR-22 simulates the effect of serotonin treatment, further increasing CPEB levels and enhancing long-term facilitation. Conversely, miR-22 overexpression downregulates CPEB and impairs long-term facilitation. Mouse CPEB-3 is also likely susceptible to miRNA regulation in neurons (Morgan et al. 2010).
CPEB aggregation is also regulated by posttranslational modification. For example, protein SUMOylation is an inhibitory constraint that regulates the aggregation and activity of CPEB-3 (Drisaldi et al. 2015). As mentioned previously, in the basal state, CPEB-3 is a repressor and is soluble. Under these circumstances, it is SUMOylated in hippocampal neurons both in vitro and in vivo. Following neuronal stimulation, CPEB-3 is converted into an active form that promotes the translation of target mRNAs, and this process is associated with a decrease of SUMOylation and an increase in aggregation. These findings suggest a model whereby SUMO regulates translation of mRNAs and structural synaptic plasticity by modulating the aggregation of a functional prion.
In addition to SUMOylation, CPEB-3 is also regulated by ubiquitination. Specifically, CPEB-3 interacts with Neuralized1, an E3 ubiquitin ligase (Pavlopoulos et al. 2011). CPEB-3 colocalizes with Neuralized1 in hippocampal neurons, whereas mice overexpressing Neuralized1 in the forebrain show heightened levels of CPEB-3 (but not other CPEBs) in the hippocampus. This interaction, which is mediated by the N-terminal, prion-like domain of CPEB, leads to the monoubiquitination and consequent activation of CPEB-3. Strikingly, overexpression of Neuralized1 mimics synaptic activity and activates CPEB-3 in cultured hippocampal neurons. In contrast, inhibition of Neuralized1 reduces the steady-state levels of monoubiquitinated CPEB-3, leading to the shortening of the poly(A) tails, reduced translation of GluA1 and GluA2 mRNAs, and, finally, interference of hippocampal-dependent memory and synaptic plasticity (Pavlopoulos et al. 2011). These results suggest a model whereby Neuralized1-mediated ubiquitination facilitates hippocampal plasticity and hippocampal-dependent memory storage by modulating the activity of CPEB-3-dependent protein synthesis and synapse formation.
Together, these examples illustrate that CPEB aggregation is regulated by a number of physiological signals and suggest that more of these mechanisms are likely to be discovered. Although it is increasingly clear that prion-like aggregation can perform a variety of positive functions in different biological contexts, the existence of multiple regulatory mechanisms strongly suggests that the detrimental effects of potential nonspecific aggregation must be counterbalanced by robust molecular and cellular feedback loops.
STRUCTURAL STUDIES OF CPEB
Unlike the case for conventional prions, Aplysia CPEB, Drosophila Orb2, and mouse CPEB-3 undergo physiologically regulated conversion from a soluble to an aggregated state. Are there structural differences between prions and prion-like proteins that account for regulated versus pathological aggregation?
In addition to β-sheet motifs that are typical of aggregation-prone proteins, ApCPEB also has coiled-coil α-helices that can mediate prion-like oligomerization (Fiumara et al. 2010), consistent with solid-state nuclear magnetic resonance (NMR) analyses showing that the prion-like domain is not solely composed of β-sheet, but also has a novel mixed structure containing helical and random coil stretches (Raveendra et al. 2013). However, unlike β-sheets, coiled-coils are responsive to environmental signals and therefore can be regulated (Fiumara et al. 2010). The propensity of the prion domain of ApCPEB to form α-helical coiled-coil structures enables the β-sheet to form a fiber-axis scaffold. In turn, this would allow the C-terminal domains to be free to bind mRNA on the surface of the fiber-axis scaffold (Raveendra et al. 2013). Such a structure would allow the coordinated translation of the population of interrelated mRNAs required for the stabilization of synaptic growth (Fig. 5).
Figure 5.
Functional prions are likely to have a distinctive regulatable structure. The coiled-coils of the prion domains are stacked together, exposed, and free to bind mRNAs on the surface of the β-sheet fiber-axis scaffold. This structure can allow the coordinated translation of the population of interrelated mRNAs required for stabilization of synaptic growth.
THE SEARCH FOR ADDITIONAL FUNCTIONAL PRION-LIKE PROTEINS IN THE BRAIN
If a single example of a functional prion in the brain is a biological curiosity, then the discovery of a second example establishes a biological principle. Toward this goal, we have identified TIA-1 as a novel functional prion-like protein in the mouse hippocampus (JB Rayman, X Li, and ER Kandel, et al., unpubl.). TIA-1 is a cellular stress response protein that facilitates the assembly of cytoplasmic aggregates known as stress granules when a cell is exposed to extreme environmental conditions (Kedersha and Anderson 2002). Stress granules are temporary sites of storage for RNAs that are not required during the stress response. By maintaining translational arrest of this population of RNAs, stress granules prioritize the expression of genes that power the restoration of homeostasis, ensuring that cellular energy is not wasted on spurious translation (Kedersha and Anderson 2002). The capacity of TIA-1 to form stress granules is highly dependent on its C-terminal prion-like domain (Gilks et al. 2004), whereas high-molecular weight, SDS-resistant aggregates of TIA-1 are detected in hippocampal extracts from mice shortly after exposure to cellular stress (JB Rayman et al., unpubl.). Furthermore, mouse TIA-1 aggregates are amyloid-rich and show heritability in a yeast assay (Li et al. 2014). It is likely that the role of TIA-1 in the brain is to protect neurons from a variety of environmental insults that, if unchecked, could lead to neurodegenerative changes. However, the notion that TIA-1 may also play a role in learning and memory is indicated by the fact that blocking prion-like aggregation of TIA-1 in Aplysia neurons impairs long-term facilitation (JB Rayman et al., unpubl.). We are also in the process of addressing the role of TIA-1 in the mouse brain. Although the latter studies are still at an early stage, these observations collectively support the notion that prion-like proteins can contribute to normal physiological processes in the nervous system.
CONCLUDING REMARKS
Prions were initially identified as the causative agents of a class of fatal neurodegenerative diseases of the mammalian brain (Prusiner 1998). However, the discovery of fungal prions, which do not cause disease, suggested that prion-like mechanisms may serve normal biological functions (Lindquist 1996). Indeed, the realization that protein conformational switches could explain stable transmission of phenotypes in yeast dates back more than 20 years (Wickner 1994). A number of fungal prions that have been identified as epigenetic determinants that alter a range of cellular processes, including metabolism and gene expression, have now been described (Tompa and Friedrich 1998; Eaglestone et al. 1999; True and Lindquist 2000; True et al. 2004).
In addition, there are now several examples of functional prion-like proteins in mammals. In this review, we describe prion-like proteins in the brain, the CPEBs, which are the earliest examples of prion-like proteins that are functional in the aggregated state. CPEB acts as a conformational switch in the brain to enable the stabilization of memory. Because of the common features of prion-like propagation of CPEB in snails, flies, and mice, along with those of other prion-like proteins in eukaryotes, we suggest that these proteins are not biological anomalies, but instead use a common structural mechanism that enables a highly sensitive, robust, and stable response to important physiological signals that could not be accomplished by other polymeric mechanisms. Thus, the preponderance of amyloid-based diseases in the nervous system may be linked to the presence of prions in the nervous system that serve normal physiological functions as long as their aggregation is regulated. It is tempting to speculate that there are additional proteins besides CPEB and TIA-1 that serve normal physiological functions in the prion state—both in the nervous system and in other cellular contexts.
ACKNOWLEDGMENTS
We are grateful for the generous support from the Howard Hughes Medical Institute and Cohen Veterans Bioscience.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Bally-Cuif L, Schatz WJ, Ho RK. 1998. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev 77: 31–47. [DOI] [PubMed] [Google Scholar]
- Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey CH, Kandel ER. 1999. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell 99: 221–237. [DOI] [PubMed] [Google Scholar]
- Chang JS, Tan L, Wolf MR, Schedl P. 2001. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 128: 3169–3177. [DOI] [PubMed] [Google Scholar]
- Drisaldi B, Colnaghi L, Fioriti L, Rao N, Myers C, Snyder AM, Metzger DJ, Tarasoff J, Konstantinov E, Fraser PE, et al. 2015. SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep 11: 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J 18: 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, et al. 2015. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 86: 1433–1448. [DOI] [PubMed] [Google Scholar]
- Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. 2010. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell 143: 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumara F, Rajasethupathy P, Antonov I, Kosmidis S, Sossin WS, Kandel ER. 2015. MicroRNA-22 gates long-term heterosynaptic plasticity in Aplysia through presynaptic regulation of CPEB and downstream targets. Cell Rep 11: 1866–1875. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wickens MP. 1989. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev 3: 2151–2162. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. 1997. Synaptic tagging and long-term potentiation. Nature 385: 533–536. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Richter JD. 1996. Mouse cytoplasmic polyadenylylation element binding protein: An evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc Natl Acad Sci 93: 14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. 2002. Translational control of the embryonic cell cycle. Cell 109: 473–483. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD. 2006. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J 25: 4865–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. 2001. The molecular biology of memory storage: A dialogue between genes and synapses. Science 294: 1030–1038. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. 2002. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Keleman K, Krüttner S, Alenius M, Dickson BJ. 2007. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci 10: 1587–1593. [DOI] [PubMed] [Google Scholar]
- Krüttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. 2012. Drosophila CPEB Orb2A mediates memory independent of its RNA-binding domain. Neuron 76: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lim CS, Park H, Lee JA, Han JH, Kim H, Cheang YH, Lee SH, Lee YS, Ko HG, et al. 2007. Nuclear translocation of CAM-associated protein activates transcription for long-term facilitation in Aplysia. Cell 129: 801–812. [DOI] [PubMed] [Google Scholar]
- Li X, Rayman JB, Kandel ER, Derkatch IL. 2014. Functional role of Tia1/Pub1 and Sup35 prion domains: Directing protein synthesis machinery to the tubulin cytoskeleton. Mol Cell 55: 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. 1996. Mad cows meet mad yeast: The prion hypothesis. Mol Psychiatry 1: 376–379. [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, et al. 2012. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148: 515–529. [DOI] [PubMed] [Google Scholar]
- Martin KC, Kandel ER. 1996. Cell adhesion molecules, CREB, and the formation of new synaptic connections. Neuron 17: 567–570. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. 1997. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell 91: 927–938. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. 1989. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev 3: 803–815. [DOI] [PubMed] [Google Scholar]
- Morgan M, Iaconcig A, Muro AF. 2010. CPEB2, CPEB3 and CPEB4 are coordinately regulated by miRNAs recognizing conserved binding sites in paralog positions of their 3′-UTRs. Nucleic Acids Res 38: 7698–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. 2011. Neuralized1 activates CPEB3: A function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147: 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. 1998. The prion diseases. Brain Pathol 8: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 47: 601–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendra BL, Siemer AB, Puthanveettil SV, Hendrickson WA, Kandel ER, McDermott AE. 2013. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol 20: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KE, Condic ML, Eisenberg LM, Yost HJ. 1999. Spatially regulated translation in embryos: Asymmetric expression of maternal Wnt-11 along the dorsal–ventral axis in Xenopus. Dev Biol 214: 288–297. [DOI] [PubMed] [Google Scholar]
- Si K, Kandel ER. 2016. The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. 2003a. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell 115: 893–904. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. 2003b. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 115: 879–891. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. 2010. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140: 421–435. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD. 1996. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J 15: 2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. 2015. The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Rep 11: 1772–1785. [DOI] [PubMed] [Google Scholar]
- Tan L, Chang JS, Costa A, Schedl P. 2001. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development 128: 1159–1169. [DOI] [PubMed] [Google Scholar]
- Theis M, Si K, Kandel ER. 2003. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci 100: 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Friedrich P. 1998. Prion proteins as memory molecules: An hypothesis. Neuroscience 86: 1037–1043. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431: 184–187. [DOI] [PubMed] [Google Scholar]
- Weissmann C. 2004. The state of the prion. Nat Rev Microbiol 2: 861–871. [DOI] [PubMed] [Google Scholar]
- Wickner RB. 1994. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]