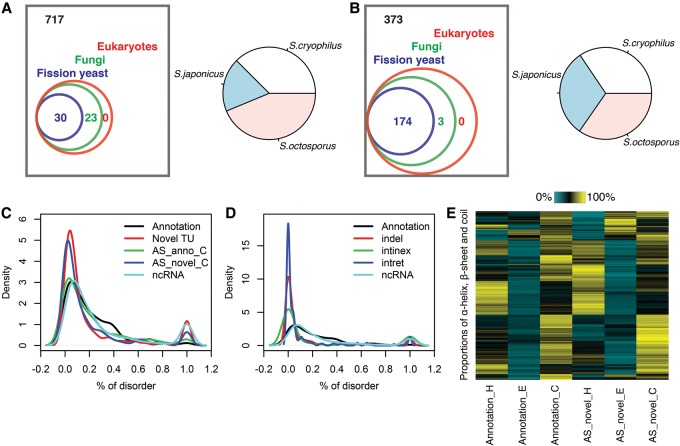
Figure 7.
Conservation and secondary structure analysis. (A) Left panel shows the protein BLAST results when using the 770 novel TUs as query sequences. Numbers of novel TUs with alignment hits in different taxa are shown, where the same TU potentially has hits in multiple taxa. Right panel shows the proportion of hits in different fission yeasts. (B) Left panel shows protein BLAST results using as query sequence only the modified part of the 550 protein sequences encoded by AS isoforms. Numbers of AS isoforms with alignment hits in different taxa are shown, where the same AS isoform potentially has hits in multiple taxa. Right panel shows the proportion of hits in different fission yeasts. (C) Distribution of percent of disorder predicted by RaptorX (Källberg et al. 2012). Five classes of protein sequences were examined: full length annotated proteins, longest proteins predicted from novel TUs, C terminus amino acid sequence starting at the first alteration in AS isoforms and its annotated counterpart, and longest proteins predicted from annotated ncRNA. (D) Distributions of percent of disorder predicted for annotated proteins, amino acids affected by insertions and deletions (indel), intron retentions (intret) and introns in exons (intinex) from AS isoforms, and annotated ncRNA. (E) Proportions of α-helix (H), β-sheet (E) and coil (C) predicted from C terminus amino acid sequence starting at the first alteration in AS isoforms and its annotated counterpart.