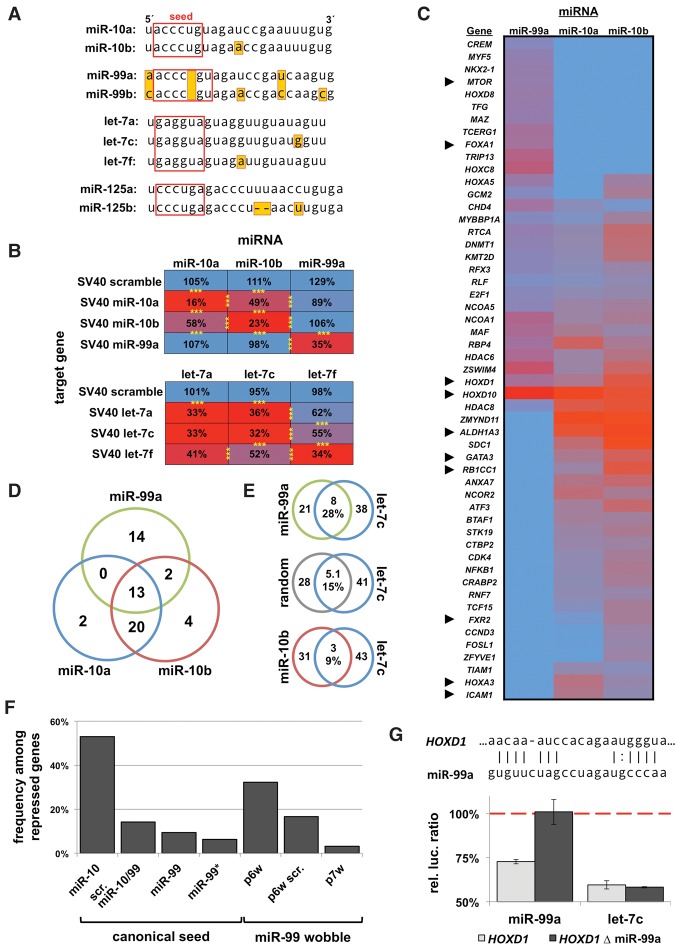
Figure 5.
miRNA family members have distinct target specificity. (A) miRNA sequences and their family members used in this study. (B) Targeting specificity of miR-10 and let-7 family members for perfectly complementary targets in the SV40 3′ UTR. Results are represented as percentage of luciferase signal normalized to a negative miRNA control. n = 4; (***) P < 0.001. (C) Results of a 3′ LIFE assay testing 375 3′ UTRs for targeting by miR-99a, miR-10a, and miR-10b (n = 5, total ∼8000 dual luciferase reactions). Heatmap of 3′ UTRs with repression (rep. <0.8, P < 0.05) by any one of the three miRNAs. Arrowheads denote genes further discussed in this study. (D) Venn diagram of genes exhibiting any degree of repression from screen in C. (E) Commonly regulated genes between genomically clustered miRNAs (miR-99a/let-7c) share more common targets than nonclustered miRNAs (miR-10/let-7c). (F) Occurrence of miR-99 seed in repressed genes. (Column 1) Frequency of miR-10b seed among genes repressed by miR-10b; (column 2) scrambled permutation of the miR-10/miR-99 seed among miR-99 hits; (column 3) canonical miR-99; (column 4) miR-99* seed; (columns 5–7) miR-99 seeds with wobbles allowed in indicated positions. (G) Deletion analysis of the p6 wobble seed motif targeted by miR-99a in the 3′ UTR of HOXD1 (nucleotides 577–599 from stop codon) rescues repression by miR-99a, but not let-7c, which also targets this 3′ UTR (n = 4).