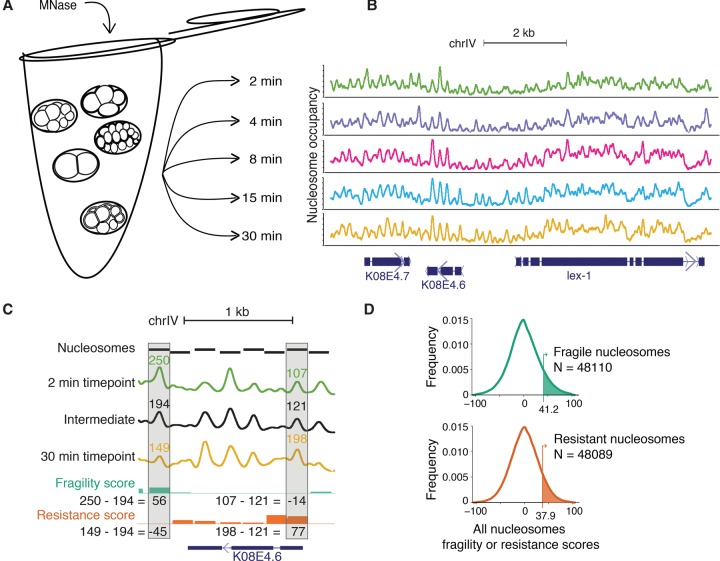
Figure 1.
An MNase digestion time-course on C. elegans embryos. (A) Mixed-stage embryos were collected from gravid hermaphrodites by bleach treatment. Dissociated nuclei from mixed-stage embryos were incubated with MNase for 2, 4, 8, 15, or 30 min. (B) Paired-end reads from each timepoint were mapped to the C. elegans genome, normalized, and Gaussian smoothed for display. High signals represent regions of the genome protected from MNase digestion. Region plotted: Chr IV position 12,074,951–12,084,347. (C) Calculation of fragility and resistance scores. (Fragility) For each nucleosome, the average occupancy of the intermediate timepoints is subtracted from the 2-min timepoint. (Resistance) For each nucleosome, the average occupancy of the intermediate timepoints is subtracted from the 30-min timepoint. Intermediate timepoints are 4, 8, and 15 min. Region plotted is Chr IV position 12,076,980–12,078,364. (D) Distribution of fragility and resistance scores at all nucleosomes. The top 10% of each class (shaded in green and orange, respectively) were considered “fragile” or “resistant.”