Abstract
Innate immune cells are endowed with many nucleic acid receptors, but the role of sequence in the detection of foreign organisms remains unclear. Can sequence patterns influence recognition? And how can we infer those patterns from sequence data? Here, we detail recent computational and experimental evidence associated with sequence-specific sensing. We review the mechanisms underlying the detection and discrimination of foreign sequences from self. We also describe quantitative approaches used to infer the stimulatory capacity of a given pathogen nucleic acid species, and the influence of sequence-specific sensing on host-pathogen coevolution, including endogenous sequences of foreign origin. Finally, we speculate how further studies of sequence-specific sensing will be useful to improve vaccine design, gene therapy and cancer treatment.
Keywords: Innate Immunity, Pattern Recognition Receptors, RNA, Sequence Patterns, Virus, Cancer
Microbial-specific Sequence Motifs are a Class of Pathogen-associated Molecular Pattern
The innate immune system detects the presence of foreign organisms and initiates a coordinated response to eliminate infectious threats. Among the microbial products sensed by innate immune effectors, efficient recognition of nucleic acids (DNA and RNA) is critical, as suggested by the existence of several families of receptors specific to these ligands (Table 1). However, the detection of nucleic acids also presents a risk for self-recognition, and self-activation in response to host nucleic acids is associated with many autoimmune diseases. Inappropriate detection of self-molecules is prevented through subcellular compartmentalization of receptors, degradation of self nucleic acids by endogenous nucleases, and specialization of innate receptors that detect conserved microbial features absent from the host. Typical microbial features recognized by the innate immune system include distinct products of post-transcriptional processing, such as nucleotide chemical modifications and specific 5′- or 3′-moieties. In addition, microbial genomes display specific nucleic acid patterns at a different frequency compared to the human genome. These variations, such as frequencies of nucleotide words, are sufficient to assign biological sequences to taxonomic spaces, at least broadly [1].
Table 1.
Interactions between viral sequence patterns and innate immune pathways deciphered by large-scale synonymous recoding of viral genomes
| Virus | Sequence pattern studied | Innate immune pathway involved | Reference |
|---|---|---|---|
| Influenza | (A/U)CG(A/U) | IFN-I secretion, TLR7-dependent | [23] |
| Simian Immunodeficiency Virus | Nucleotide bias | IFN-I, IRF3-dependent | [28] |
| Echovirus | CpG and UpA | Unknown PRR, IRF3-independent | [24] |
| Infuenza | CpG and UpA | Pro-inflammatory cytokines Unknown PRR |
[25]. |
| Foot and Mouth Disease Virus | Codon pair bias | IFN-I Pro-inflammatory cytokines |
[26] |
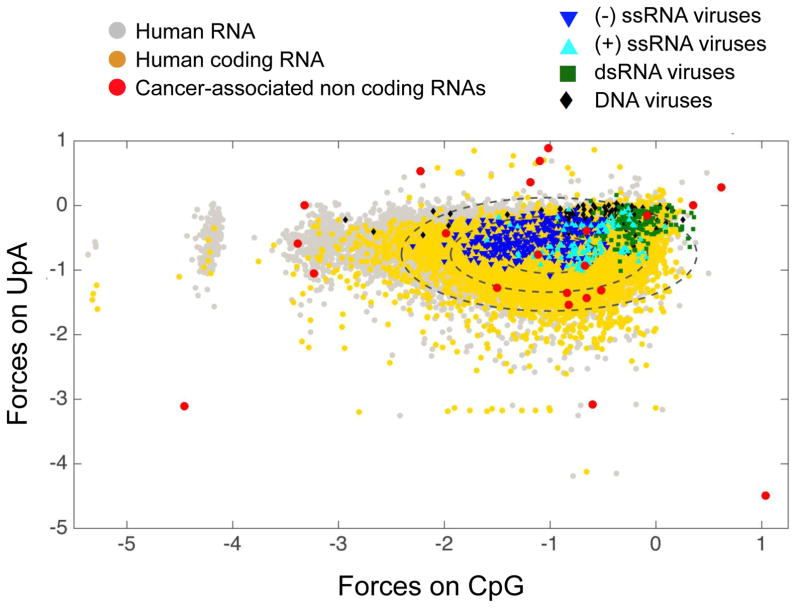
As a result, coding and non-coding transcripts pertaining to a single “self” genome display specific sequence properties that can be used to distinguish them computationally from a “non-self”, foreign, genome (Fig. 1) [2]. For example, differential CpG and UpA usage patterns create a metric that computationally segregates the genomes of some human viruses, including positive-, negative-, and double-stranded RNA viruses, DNA viruses and, interestingly, many transcripts only found in high abundance in tumors, into taxonomic spaces that are distinct from the typical human transcriptome (Fig. 1). Thus, differences in sequence patterns theoretically can be used by host cells to distinguish self sequences from non-self.
Figure 1. CpG and UpA Dinucleotide Distributions Among Different Classes of Transcripts.
The distribution of forces on CpG and UpA dinucleotides among non-coding and coding RNA allows one to define a landscape of human transcripts (gray dots). Forces represent the entropy penalty for non-random motif usage, with positive forces indicating over-representation and negative under-representation of the motif. Among this landscape, a particular space is occupied by human coding sequences (yellow dots, each ellipse indicates 1 SD from the mean, 95% of transcripts are distributed inside the median ellipse). The same metric applied to human viruses shows that viral genomes occupy a restricted space, yet most human viruses mimic their human host to within two standard deviations, with a set of exceptions, particularly among the dsRNA viruses. Interestingly, several non-coding RNA upregulated in pancreatic and other cancer cells identified in [77] are clear outliers from normal CpG and UpA use (red dots). This figure is an agglomeration of sequences and analyses from [5, 76, 77].
Here, we describe both the computational approaches to inferring a foreign sequence and the sequence specificity of receptors involved in innate sensing of nucleic acids. We discuss the newly described importance of sequence-specific sensing on the immunostimulatory properties of endogenous elements of foreign origin, along with other mobile elements and non-coding RNAs, and their emerging role in cancer immunity.
Computational Approaches to Define a Foreign Sequence
The earliest computational approaches to defining a foreign sequence used motif usage as a self versus non-self discriminant. A pioneering study showed that a metric based on dinucleotide usage could adequately discriminate a random contiguous segment of human DNA from a random contiguous segment of bacterial DNA [3]. The primary dinucleotide that enforces this discrimination is CpG. Indeed, CpG methylation of DNA, a feature vastly more prevalent in mammals than bacteria, promotes cytosine deamination, leading to accelerated cytosine to thymine mutation. Thus, CpG methylation is thought to account for underrepresentation of CpG motifs in mammalian genomes.
A few years earlier, it was found that a pattern recognition receptor (PRR), Toll-like receptor 9 (TLR9) senses DNA containing unmethylated CpGs in the endosomes of mammalian cells [4]. Hence, the same pattern detected computationally to discriminate mammalian and bacterial genomes is associated with an actual innate receptor performing the same task. Subsequently, new methods were introduced to computationally identify such discriminatory motifs. One approach introduced further constraints on genomes, fixing not just nucleotide frequency but also the coding sequence of a virus and its codon usage. By randomizing the genome sequence while fixing these constraints, it is possible to identify patterns that discriminate pathogen and host genomes, independently of their effect on the codon usage or amino acid sequence [5].
The more constraints introduced, the more computationally taxing randomization becomes. These approaches thus become ill suited to longer genomes or whole transcriptome analyses, where the size of the dataset requires more efficient methods. Motivated by this issue, the problem was recently recast using methods from statistical physics, which avoid tedious genomic randomizations to find atypical patterns [6]. The computational speed associated with this improved conceptual language allows larger datasets, such as eukaryotic transcriptomes, to be efficiently analyzed. In this framework, one defines a force on a genomic pattern as the information entropy cost for a genome to have that pattern at the observed frequency. In other words, a pathogen pays a measurable evolutionary cost to reorganize the information in its genome in a non-random way in order due to the influence of specific sequence patterns targeted by a PRR, reaching an equilibrium with its host that balances between the selective forces on the pattern and randomizing entropic forces. As a consequence when a pathogen changes hosts, potentially encountering a new set of PRRs, it will evolve towards a new equilibrium between PRRs and sequence entropy.
Microbial Avoidance of Sequence Patterns Suggest Immunostimulatory Properties
The first demonstration of sequence-specific immune sensing of foreign nucleic acids came from the discovery that CpG motifs in bacterial DNA trigger B-cell activation [4]. The same immunostimulatory potential of certain dinucleotides was suggested by genomic analyses that identified a pervasive suppression of CpG and UpA dinucleotides in RNA viruses infecting mammals [7] [8]. Later, an evolutionary survey of influenza A upon transition from avian to human hosts suggested a selection pressure against CpG dinucleotides in A/U rich contexts [5]. Interestingly, this motif pattern is highly under-represented in the sequences of several human innate immune genes, such as the type I interferon (IFN-I) family. These RNA, expressed at high quantity almost exclusively during the response to viral infection, may have evolved to avoid further immune stimulation, which could create a positive feedback loop [9]. Similarly, in a study comparing the dinucleotide usage patterns of Flaviviridae viruses infecting vertebrate or invertebrate organisms, the vertebrate-infecting viruses display a decreased CpG frequency not observed in insect-only viruses [10].
Nucleotide word usage is not the only sequence feature that has been associated with sequence-specific immune detection. Yet another feature comprises large secondary RNA structures observed in several RNA virus families. Their presence has been linked to viral fitness, in vivo persistence and escape of innate immune detection [11]. Furthermore, as for many viral families, lentiviruses, such as Human immunodeficiency virus (HIV), possess a genome with a significant nucleotide ratio bias compared to human genes [12]. One hypothesis explaining this biased composition is that, as a retrovirus, HIV is subject to innate pressure from RNA editing enzymes such as APOBEC3G, which can edit cytosine nucleic acids in a motif specific fashion, inducing a mutational bias [13]. Likewise, an analysis of different HIV-1 virus subtypes has uncovered a correlation between HIV-1 nucleotide bias, its ability to elicit an innate immune response, and its pathogenicity [14]. Thus, while there is evidence that some specific immunostimulatory motifs are being selected against, microbial sequences also evolve under both intrinsic and host-related constraints, leading to some pattern usage biases not necessarily identical to those of their hosts.
Large-scale Synonymous Recoding of Viral Genomes Indicates Innate Detection of Specific Sequences
A popular approach for studying the interaction of the immune system with viral sequence patterns is the large-scale synonymous recoding of viral genomes [15]. Taking advantage of the genetic code’s degeneracy, this strategy consists of replacing whole portions of viral genomes with sequences either enriched for or depleted of the pattern of interest, without altering the protein coding sequence. Pioneering studies introduced rare codons in the capsid-coding region of poliovirus genome and observed that it led to viruses with altered replication fitness, lower virulence and infectivity [16] [17]. Similar strategies, including alterations in codon usage, codon pair usage, and/or dinucleotide content also led to viral attenuation, the mechanisms of which are currently under investigation. Depending on the studies, authors have thus far reported inhibition of protein synthesis [17] [18] [19] [20], inhibition of viral cDNA synthesis [21] or reduction of virion infectivity [17] [22]. Interestingly, several of these investigations, listed in Table 1, found that modification of viral genomes modulates their interactions with the innate immune system. Of note, a team generated two non-replicative influenza virus mutants in which the viral PB1 polymerase gene was replaced by a green fluorescent protein (GFP) gene. One virus contained a GFP sequence with 21 (A/U)CG(A/U) motifs, while the other contained only 2. Upon infection of human primary plasmacytoid dendritic cells, the mutant with more CpG motifs elicited higher levels of interferon-α [23].
The immune impact of modifying CpG and UpA dinucleotide frequencies in viral genomes was later confirmed by additional studies. In one study, increasing frequency of CpG and UpA dinucleotides in the genome of the picornavirus echovirus 7 produced mutants with impaired replication kinetics. The authors found no evidence for differential recognition of the virus by an already known PRR. However, experimental use of kinase inhibitors reverted the attenuation phenotype of the mutants, suggesting a role for an unknown PRR in mediating a dinucleotide usage-specific immune response [24]. In a follow-up study on influenza virus, the authors demonstrated that, despite marked attenuation of replication, virus mutants enriched in CpG and UpA induced higher inflammatory cytokines and adaptive responses in vivo [25].
Another team generated a replicative mutant of Foot-and-Mouth Disease Virus, by replacing codons pairs by synonymous codon pairs underrepresented relative to the human genome. The mutant exhibited an increase in its overall CpG content resulting in attenuated replication in vitro and higher levels of type I interferon, IFN-stimulated genes (ISG) and proinflammatory cytokines [26]. A different team produced a mutant of simian immunodeficiency virus (SIV) with optimized nucleotide composition by lowering nucleotide bias in the pol gene, resulting in increased CpG frequency, and overall reduction of dinucleotide bias [27]. Optimized SIV mutant displayed normal replication kinetics but a decreased ability to induce IFN-I expression. Moreover, IFN regulatory factor 3 (IRF3) was required for IFN-I induction, suggesting a role for cytosolic sensors in detecting SIV biased RNA [28].
Sequence Specificity of Known Human Receptors
Several nucleic acid sensors have been proposed or suspected to sense specific microbial sequences, as listed in Table 2. We categorize these receptors depending on two aspects. The first is subcellular localization, since one would hypothesize that sensitivity of PRRs strongly depends on spatial proximity to self-derived ligands. According to this hypothesis, nucleic acid receptors located in endosomes and cytoplasmic DNA sensors should not bind ligands in a highly sequence-dependent manner, and, cytoplasmic RNA PRRs would have developed higher specificity for foreign sequences. This does not always hold true. The second aspect is their sequence specificity and the techniques used to infer them.
Table 2. Known receptors that sense specific nucleic acid sequences.
We listed each receptor according to the nature of its ligand, the sequence it recognizes, its subcellular localization and the methods used to determine the sequence specificity. NGS: Next Generation Sequencing. EMSA: Electrophoretic Mobility Shift Assay.
| Receptor | Sequence/pattern | Method of determination | Localization | Reference | |
|---|---|---|---|---|---|
| RNA | RIG-I | AU-rich sequence and dsRNA structures | NGS and crystal structure | Cytosol | [41] [42] [43] [48] [49] [50] |
| MDA5 | AU-rich sequence and dsRNA structures | NGS and crystal structure | Cytosol | [48] [49] [51] | |
| DDX17 | CA- and CT-repeat, (GTA)CATCC(CTA) motif and stem-loop structure | NGS | Cytosol and Nucleus | [53] [54] | |
| IFIT 2 | AU-rich ssRNA sequence | Crystal structure and EMSA | Cytosol | [52] | |
| TLR7 | U(C/G)U or U(U/A)N and (A/U)CG(A/U) motifs. G residues and U-rich ssRNA |
Cytokine measure after stimulation | Endosome | [23] [32] [34] | |
| TLR8 | U-rich ssRNA degraded into U residues and short oligonucleotides | Crystal Structure | Endosome | [33] | |
| TLR13 | GAAAGACC and stem- loop-like structure | Crystal Structure | Endosome (mouse) | [81] | |
| DNA | TLR9 | RRCGYY | Crystal structure | Endosome | [38] |
| cGAS | Short dsDNA flanked with at least 3 G | Cytokine measure after stimulation + receptor binding assay | Cytosol | [57] | |
| Sox2 | (C/T)(A/T)TTGT(T/C)ATG CAAAT | Cytokine measure after stimulation + receptor binding assay | Cytosol | [58] |
Nucleic Acid-sensing Endosomal TLRs
TLR3, TLR7, TLR8, TLR9 and TLR13 (Box 1) are endosomal receptors that sense nucleic acids that have been endocytosed or phagocytosed. The crystal structures of most TLRs bound to their respective ligands have been deciphered. TLRs have a ligand-recognition domain facing the lumen of the endosome, a transmembrane domain, and a signaling domain facing the cytosol. Despite compartmentalization, TLR7, TLR8 and TLR9, but not TLR3, demonstrate a certain level of sequence specificity.
Box 1. The Intriguing Specificity of Mouse TLR13.
TLR13 is an endosomal TLR expressed in mice but not humans. TLR13 recognizes its ligand in a stringently sequence-specific manner, sensing a highly conserved bacterial 23S rRNA sequence that contains 5′-GAAAGACC-3′ [82] [83]. Notably, this sequence is found within a region of RNA targeted by certain antibiotics, and clinical isolates of Staphylococcus aureus resistant to these antibiotics are unable to stimulate mouse TLR13 [83]. The structure of mouse TLR13 bound to a 13-nt ssRNA derived from 23S rRNA has also been characterized. When bound to TLR13, the ssRNA folds into a stem-loop-like structure that is different from its usual shape in the bacterial ribosome, and that is also required for activation of TLR13. However, most of the ssRNA nucleotides make base-specific contacts with the surface of TLR13, accounting for its sequence specify [81]. Interestingly, a viral-derived 16-nt ssRNA, containing the related sequence GAAAGACU and predicted to form a similar stem-loop-like structure also induces TLR13 activation [81]. Overall, TLR13 functions as a sequence- and conformation-specific PRR.
TLR7 and TLR8 recognize ssRNA and share a general specificity for U-rich ssRNA [29] [30]. A study showed that a uridine-rich tetramer is the minimal sequence required to elicit TLR7 and TLR8 responses and further suggested TLR8 has increased specificity for AU-rich sequences, while TLR7 has a comparatively heightened specificity for GU-rich sequences [31]. A second investigation found that endogenous nucleic acid sequences induced a level of TLR7 activation in murine B cells directly proportional to their uridine-content, and, particularly, to the frequency of two motifs, U(C/G)U and U(U/A)N [32]. The crystal structure of TLR8 in complex with ssRNAs showed that the receptor contains two ligand-binding sites and that synergistic binding of both sites to their respective ligand is required for signal transduction. The first site binds a uridine residue and the second binds a short guanosine-containing oligoribonucleotide, like UG [33]. After crystallization, intact ssRNA was not observed bound to the TLR8 structure, suggesting TLR8 binds products of ssRNA degradation catalyzed by yet unknown nucleases and phosphatases [33]. A follow-up study performed on TLR7 showed that it also contains two ligand-binding sites. Indeed, TLR7 acts as a dual receptor that binds both guanosine and uridine-containing ssRNA. Interestingly, the ssRNA-binding site is spatially and structurally distinct form that of TLR8. However, as for TLR8, synergistic binding of both ligands is required for efficient activation [34]. Interestingly, use of guanosine derivatives such as Resiquimod (R848) shows chemicals with a minimal nucleotide structure can act as agonist of both TLR7 and TLR8 [35].
TLR9 recognizes bacterial and viral ssDNA containing an unmethylated CpG motif with a strong level of sequence specificity. Indeed, depending on its sequence, a DNA strand will act either as TLR9 agonist or antagonist [36]. The TLR9 immunostimulatory CpG motif, which consists of a hexamer with a central unmethylated CpG, was first described as RRCGYY, with R representing a purine and Y a pyrimidine [4]. In humans, the CpG motif with formula GTCGTT has been proposed as an optimal ligand [37]. The crystal structure of TLR9 has been characterized. Using agonist 12-mer oligonucleotides, it was found that the consensus hexamer sequence, and no other part of DNA, is directly recognized by TLR9 [38]. TLR9’s specificity for CpG motifs and their flanking sequences is determined by precise van der Waals’ interactions and water-mediated hydrogen bonds between multiple TLR9 amino acids and DNA bases. Of note, TLR9 recognizes CpG in DNA sequence only. However, CpG dinucleotide are also suppressed in many RNA viruses, which do not produce DNA at any step of their replication cycle. Thus, it’s likely that TLR9 specificity drives CpG suppression in DNA viruses for example, but is unlikely to be involved in ssRNA and dsRNA viruses genome shaping. Additionally, the backbone phosphate is needed for recognition. In another study, TLR9 has been shown to recognize DNA:RNA hybrids [39]. Curiously, ssDNA fragments isolated from these hybrids were not potent in activating TLR9, presumably due to the absence of CpG motifs. Overall, these results suggest an intriguing dependency of TLR9 sequence specificity to the nature of its ligand.
RIG-I-like Receptors
RIG-I
The best-studied cytosolic RNA sensors are retinoic acid-inducible gene I (RIG-I) and melanoma differentiation factor 5 (MDA5). RIG-I primarily relies on structural features like 5′ triphosphate extremities and dsRNA fragments to detect viral RNA [40]. Despite the high-resolution data of crystal studies, a consensus on the definition of a RIG-I ligand has yet to be reached [41] [42] [43]. RIG-I has been shown to recognize specific sequences of viral genomes, such as poly-U/UC motifs found in the 3′ untranslated region of hepatitis C virus and in the N gene of Hantaan virus [44] [45]. Several studies have also demonstrated that manipulation of RNA ligand sequences, and notably the number of uridine residues, modulate activation [46] [47]. Moreover, precipitation of cellular proteins bound to viral RNA during infection suggests that RIG-I preferentially associates with AU-rich RNAs derived from viral genomes [48] [49] [50]. As expected from its crystal structure determination, RIG-I additionally favors sequences that can form dsRNA structures, such as the sequence in the 3′-UTR of Chikungunya virus and in sequences of RNAs found in Measles, Sendai or Influenza virus defective interfering particles [48] [49] [50].
MDA5
MDA5 binds dsRNA independently of its terminal moieties and, according to structural studies, without any direct sequence specificity. Indeed, MDA5 interacts primarily with the phosphate backbone and 2′ hydroxyl groups of ribose in dsRNA, using it as a platform to stack along dsRNA in a head-to-tail arrangement. The cooperative binding of MDA5 on longer dsRNA increases its affinity, eventually leading to the proper activation of downstream signaling pathways [51]. However, next-generation sequencing (NGS) studies performed in the context of viral infection have revealed that, like RIG-I, MDA5 associates preferentially with AU-rich viral sequences [48] [49].
Other Cytosolic Sensors
IFIT2
Interferon-induced proteins with tetratricopeptide repeats 2 (IFIT2) can directly bind viral RNAs and exert antiviral functions such as inhibition of viral translation and activation of anti-viral signaling pathways. Crystal structure analysis coupled to an electrophoretic mobility shift assay indicates IFIT2 possesses a preference for AU-rich RNAs [52].
DDX17
DEAD-Box Helicase 17 (DDX17) is a multifunctional helicase that binds stem-loop structures of viral RNA in the cytosol of infected cells [53]. In the nucleus, DDX17 binds both CA- and CT-repeat elements found in mature cellular mRNAs and the (GTA)CATCC(CTA) motif found in miRNAs [54]. Thus, DDX17 uses both primary sequence and secondary structure for optimal binding to ligands (see also Box 2).
Box 2. Sequence and Structure: Patterns Inherently Intertwined.
The structure of nucleic acids depends on base-pairing interactions, and therefore is inherently linked to sequence. For instance, GC-rich RNAs tend to fold into more stable structures than AU-rich RNAs, simply because a GC pair has three hydrogen bonds whereas an AU pair only has two. Thus, GC-rich sequences may form robust secondary structures whereas AU-rich fragments should have higher flexibility [84]. In viral genomes in particular, studies have specifically correlated nucleotide composition with secondary structure. In lentiviruses, the preference for adenosine that characterizes this family is increased in single-stranded domains, but absent in double-stranded domains [12]. Similar observations have been made for Flaviviridae [85] and Coronaviridae [86].
This link between the sequence and structure bears significance for several PRRs, which depend on molecular recognition of RNA structures for immune sensing. This is the case for TLR3, which was initially identified as a dsRNA receptor [87]. It has been shown that dsRNA must meet a minimum length of 40 base pairs to activate TLR3 [88]. The crystal structure of human TLR3 complexed to dsRNA demonstrates that TLR3 recognizes the phosphate backbone of dsRNA, but makes minimal contacts with bases, confirming sequence-independent sensing [89]. Yet, several reports have since suggested RNA structures other than perfect stretches of dsRNA activate TLR3, including structures formed by bacterial and host ssRNAs [90]. These studies indicate that specific sequences may form self-RNA duplexes recognized by TLR3. Similarly, Protein Kinase R (PKR) is a PRR activated by several distinct RNA secondary structures including dsRNA, short stem-loop RNAs flanked by single-stranded tails, and misfolded RNA with structural defects, like the bulges and internal loops often found in microbial transcripts [91].
The study of immunostimulatory structures has been limited by technical challenges to determining the secondary and tertiary structures of long RNA molecules in vivo. Recently, the development of several high-throughput techniques has enabled more accurate predictions on how RNA folds inside the cell (Reviewed in [92]). These predictions propose that, contrary to microbial RNAs, cellular self-RNAs are less folded than primarily thought, strengthening the idea that PRRs discriminate pathogens based on structures adopted by their nucleic acids.
cGAS
Crystal studies show that cyclic GMP–AMP synthase (cGAS) senses cytosolic dsDNA predominantly by binding to its sugar–phosphate backbone, suggesting sequence-independent innate sensing [55] [56]. In contrast, additional work on DNA structures observed in early reverse transcripts of HIV-1 as well as endogenous retroviral elements, such as the SL2 stem loop located in the 5′ extremity of HIV’s genome, demonstrated ssDNA stem-loop structures flanked by unpaired guanosines activate cGAS in a sequence-dependent manner [57].
Sox2
Sox2 is a transcription factor that has also been shown to act as a cytosolic dsDNA receptor. To detect microbial DNA, Sox2 binds to sequence motifs in bacterial genomes such as L. monocytogenes. Interestingly, these sequences are similar to the endogenous DNA motifs Sox2 binds when acting as a transcription factor [58].
Sequence Patterns and RNA Decay
Molecular stability is another property linked to nucleic acid sequence (see also Box 3). Unique sequence elements found at the 3′-end of cellular mRNAs can mediate their degradation, such as AU-rich elements (AREs), that are characterized by a tandem repeat AUUUA sequence or a simple U-rich region [59]. AREs are observed in the sequence of many mRNAs related to the immune response and binds specific proteins, such as AU-binding factor 1 (AUF1), which recognizes AREs and targets mRNAs for rapid degradation. [60]. Interestingly, AUF1 can also directly target regions of viral RNA and inhibit viral replication, as observed during enterovirus and human rhinovirus infection [61].
Box 3. Sensing the Codon Bias?
In addition to nucleotide and dinucleotide bias, most human pathogens display a strong codon bias compared to average human codon usage. A study recently performed in yeast shows that the DEAD-box protein Dhh1p (an ortholog of human helicase Ddx6) is able to sense codon bias in cellular mRNA and induce a selective decapping and decay of biased mRNA [93]. The authors propose a mechanism where Dhh1p preferentially binds to mRNAs with high codon bias, owing to an additional interaction of Dhh1p with ribosomes, which are known to stall and accumulate during the translation of non-optimal codons stretches. According to the authors of this study, Dhh1p would thus be able to sense ribosomal speed and induce the selective degradation of codon-biased mRNA. These results echo back an older study showing that Schlafen 11, an interferon stimulated gene, could restrict retroviral infection in a codon-usage-dependent manner, this time by modulating the cellular tRNA levels during HIV infection [94]. Given the strong involvement of DEAD-box proteins in innate immunity and their ability to interact with several innate immune adaptors [95], analyzing the role of Ddx6 and its homologs in sensing microbial sequences may unravel interesting mechanisms linking codon adaptation and pathogen sensing.
Another example of a cellular intrinsic defense mechanism preventing translation of foreign genetic information is the zinc-finger antiviral protein (ZAP). ZAP binds specifically to viral RNAs containing a ZAP responsive element (ZRE) and subsequently recruits cellular RNA decay machinery [62]. The exact nature of ZRE remains to be identified. However, the resolution of the crystal structure of the ZAP RNA binding domain suggests target RNAs adopt a tertiary structure, with certain nucleotides positioned to fit into a three-dimensional cleft formed by ZAP. Interestingly, the authors suggest that the target nucleotides may not be found in a consecutive linear sequence but rather from different regions of the target RNA, accounting for difficulty in identifying a common ZRE motif [63].
Finally, the well-studied ribonuclease L (RNaseL) bridges RNA degradation and innate activation. During viral infection, RNAse L degrades host and viral RNA, preventing viral replication. Interestingly, the degradation products of RNAse L serve as RIG-I ligands, generating an IFN-I response [64]. A structural analysis suggest that RNase L recognizes the pattern UN^N, and cleaves 3′ of UN sequences [65].
Endogenous Silenced Elements: Sequences and Consequences
Overall, sequences of nucleic acids influence their structure, stability and recognition by cellular receptors. Interestingly, many human sequences, which are not transcribed under homeostatic conditions, contain sequence patterns not observed in the rest of human transcriptome (Fig. 1). Recent analysis of their interaction with the innate immune systems has unraveled unexpected new roles for these silenced elements.
During the evolution, sequences of foreign origin, predominantly stemming from retroviruses, have invaded and colonized the human germ line. Once integrated in host DNA, these sequences may have amplified their copy numbers through rounds of reinfection until eventual fixation [66]. Some retroelements, particularly recent integrations, have retained viral characteristics, like sequence-specific features of viral genomes. Under homeostatic conditions, many of these elements are transcriptionally repressed, likely preventing auto-immunity. This involves primarily epigenetic mechanisms such as histone modification and DNA methylation [66]. However, rupture of cellular homeostasis could lead to reactivation, and evidence that this process leads to the transcription of immunostimulatory ligands is beginning to surface (Fig. 2).
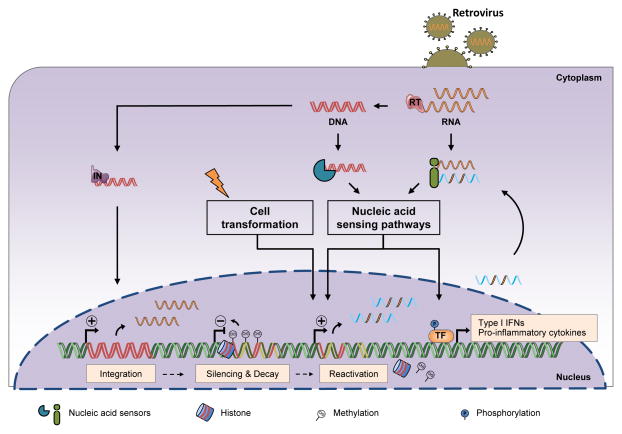
Figure 2. A Role for Microbial Sensing Pathways in Detecting Neoplasia.
During episodes of ancient infection, sequences of viral origin have been integrated into the cellular genome and subjected to active epigenetic silencing. This generally coincides with mutational decay that induces functional inactivation. However, certain sequence-specific characteristics of their viral origin may have been maintained over evolutionary time. Cell transformation or immune activation results in the transcriptional reactivation of these sequences. They may then engage receptors of the nucleic acid sensing pathways, leading to the initiation of an antimicrobial innate immune response such as production of IFN-I and pro-inflammatory cytokines. RT: Reverse-Transcriptase. IN: Integrase.
Reactivation of Transposable Elements Is Triggered and Sensed by Innate Immunity
Several studies have shown that the induction of endogenous retroelement transcription activates innate immune pathways. Reactivation of these normally silenced elements has been associated to autoimmune diseases [67]. Studies on Aicardi–Goutières syndrome (AGS) have linked the excessive IFN-I response characteristic of this condition to mutations in nucleases such as TREX1 and RNAseH. These mutations may result in inappropriate accumulation of nucleic acids and subsequent activation of RNA and DNA sensors such as MDA5 and cGAS [68] [69]. In the mouse, artificial activation of Long Interspersed Element-1 (LINE-1) has been shown to increase the expression of IFN-I and ISGs [70]. Interestingly, silenced endogenous sequences can be activated by innate immune responses, suggesting a role for these sequences in signal amplification. For instance, exposure to environmental microbes causes global modulation of endogenous retroelement transcription [71]. Moreover, treatment with IFN-I induces transcription of the short interspersed element (SINE), Alu, which in turn stimulates the secretion of proinflammatory cytokines in a TLR7-dependent manner. The authors of this study were able to identify a specific Alu sequence motif, normally bound to RNA binding protein Ro60, as immunostimulatory [72]. Similarly, activation of the B cell response with T cell-independent type 2 antigens induces transcription of endogenous retrovirus (ERV) RNAs, which are then detected via DNA and RNA sensing pathways [73]. Interestingly, this mechanism can be hijacked by viral infections. Of note, Herpes virus infection leads to the expression of SINE elements, enhancing viral replication and gene expression through activation of the antiviral NF-κB pathway [74]. Thus microbial activation of retroelement expression may function to further alert and amplify the presence of an invading pathogen.
Transcription of Silenced Sequences in Cancer
In addition to infection, the innate immune response can be triggered through expression of endogenous retroelements in tumors. Cell transformation coincides with complex, genome-wide alteration of the epigenetic landscape [75]. Treatment of certain tumor cell lines with inhibitors of DNA methyltransferase induces ERV demethylation and transcription, which correlates with an elevated IFN-I response dependent on RNA-sensing pathways [76] [77]. One is left to wonder how these endogenous retroelements sequences are distinguished from self. Although some of them can differ in sequence from self-RNA (Fig. 1), the connection of specific viral sequence patterns embedded in retroelements to their observed immunostimulatory activity has yet to be established. One particular subclass of pericentromeric repeats, the human repeat human satellite repeat II (HSATII), is greatly overexpressed in certain cancers [78]. It has been shown that HSATII sequences are enriched in the motif pattern of CpG dinucleotides in AU-rich contexts, whose detection induces the production of proinflammatory cytokines [79]. In addition to their atypical dinucleotide composition, pericentromeric repeats display other similarities with retroviral RNAs, including their ability to be reverse-transcribed [80]. Altogether, these results strengthen the proposition that endogenous retroelements bear functional roles in initiating immune responses during cancer development.
Concluding Remarks and Future Perspectives
The mammalian immune system relies on a large array of nucleic acid sensors. In recent years, increasingly refined computational studies of host and microbial genomes have greatly improved our understanding of the features that allow the distinction of self and non-self sequences. In parallel, biochemical and structural analyses of nucleic acid sensors have unraveled various degrees of sequence specificity.
Here, we reviewed both computational and experimental evidence that innate immunity restricts sequence landscapes by targeting specific sequences and sequence patterns primarily found in pathogens. These advances will likely be critical for numerous applications, including the design of optimal nucleic acid adjuvants for use in vaccines. Conversely, only a complete understanding of what constitutes a homoeostatic human transcriptome will allow the design of nucleic acid molecules devoid of immunostimulatory capacity. These should be required for gene therapy and mRNA therapy treatments. Further, we can envision the synthesis of nucleic acid sequences tailored to elicit a specific innate immune response, in order to treat particular conditions in immunotherapy (see Outstanding Questions).
Outstanding Questions Box.
Can we establish a comprehensive model of the human genome and its transcriptional landscape, sufficient enough to define a “self” -and by exclusion- a “non-self” sequence profile? Under what conditions can that landscape change?
Are there any additional, unknown innate sensing pathways specialized in sensing foreign sequences? If yes, what are the mechanisms?
What prevents microbial evolutionary evasion from specific sequences and sequence patterns? Are these constraints one can target?
Are there evolutionary advantages to maintain or enhance the properties of immunostimulatory sequences in the human genome?
What epigenetic mechanisms control the expression of immunostimulatory sequences? Can we define an epigenetic signature associated with their transcription?
Can the expression of immunostimulatory sequences be associated with specific immune markers? For instance, activation of dedicated innate immune pathways and secretion of specific cytokines?
What are the beneficial contributions of endogenous retroelements in immune activation against infection and cancer development? And what is the clinical outcome associated with their expression?
Finally, it is important to note that some host sequences derived from ancient retroviral infections have maintained characteristics of non-self sequences. Their expression, normally silenced, can be reactivated during inflammation and neoplasia, and may potentiate inflammatory responses. For these reasons, expression of foreign sequence patterns by human cells represents a marker that warrants monitoring. Further work will be required to confidently define what constitutes the landscape of “self” human sequences.
Trends Box.
Computational analyses of host and pathogen genomes lead to the identification of unique immunostimulatory sequence patterns. In parallel, structural and biochemical analyses of innate receptors confirmed the existence of specificity for unique sequence motifs.
Large-scale recoding of viral genomes uncovers new interactions between the innate immune system and sequence patterns found in viral genomes.
The sequence of nucleic acids modulates their structure, stability and direct recognition by cellular pattern recognition receptors.
Endogenous sequences of foreign origins that are normally silenced can induce innate responses through nucleic acid sensing pathways.
A better characterization of the innate pathways responsible for sensing foreign sequences will improve computational detection and bring critical insights for gene therapy and vaccine design.
Acknowledgments
NV is grateful to Dr. A. Lepelley and C. Melegari for helpful discussion and critical reading of the manuscript. NB would like to acknowledge support from the National Institutes of Health 1P30 CA 196521-01, 1R01 CA180913-01, R01 CA180913-01 and the Melanoma Research Alliance. BDG would like to acknowledge support from the National Institutes of Health P01CA087497-1, Stand Up to Cancer, the V Foundation, the Lustgarten Foundation and the National Science Foundation 1545935, along with conversations with Simona Cocco, Arnold Levine, Remi Monasson, Vladimir Roudko, and Petr Sulc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
List of references
- 1.Coutinho TJ, et al. Homology-independent metrics for comparative genomics. Computational and structural biotechnology journal. 2015;13:352–357. doi: 10.1016/j.csbj.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulveling D, et al. Identification of a dinucleotide signature that discriminates coding from non-coding long RNAs. Frontiers in genetics. 2014;5:316. doi: 10.3389/fgene.2014.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell A, et al. Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9184–9189. doi: 10.1073/pnas.96.16.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krieg AM, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 5.Greenbaum BD, et al. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS pathogens. 2008;4:e1000079. doi: 10.1371/journal.ppat.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenbaum BD, et al. Quantitative theory of entropic forces acting on constrained nucleotide sequences applied to viruses. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5054–5059. doi: 10.1073/pnas.1402285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlin S, et al. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? Journal of virology. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rima BK, McFerran NV. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. The Journal of general virology. 1997;78(Pt 11):2859–2870. doi: 10.1099/0022-1317-78-11-2859. [DOI] [PubMed] [Google Scholar]
- 9.Greenbaum BD, et al. Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PloS one. 2009;4:e5969. doi: 10.1371/journal.pone.0005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobo FP, et al. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PloS one. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witteveldt J, et al. The influence of viral RNA secondary structure on interactions with innate host cell defences. Nucleic acids research. 2014;42:3314–3329. doi: 10.1093/nar/gkt1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hemert F, et al. On the nucleotide composition and structure of retroviral RNA genomes. Virus research. 2014;193:16–23. doi: 10.1016/j.virusres.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Kim EY, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS pathogens. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vabret N, et al. The biased nucleotide composition of HIV-1 triggers type I interferon response and correlates with subtype D increased pathogenicity. PloS one. 2012;7:e33502. doi: 10.1371/journal.pone.0033502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez MA, et al. Synonymous Virus Genome Recoding as a Tool to Impact Viral Fitness. Trends in microbiology. 2016;24:134–147. doi: 10.1016/j.tim.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Burns CC, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. Journal of virology. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller S, et al. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. Journal of virology. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martrus G, et al. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology. 2013;10:78. doi: 10.1186/1742-4690-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nature biotechnology. 2010;28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keating CP, et al. The A-rich RNA sequences of HIV-1 pol are important for the synthesis of viral cDNA. Nucleic acids research. 2009;37:945–956. doi: 10.1093/nar/gkn1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tulloch F, et al. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife. 2014;3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Baranda S, et al. Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. Journal of virology. 2011;85:3893–3904. doi: 10.1128/JVI.01908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atkinson NJ, et al. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic acids research. 2014;42:4527–4545. doi: 10.1093/nar/gku075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaunt E, et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. eLife. 2016;5:e12735. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-San Segundo F, et al. Synonymous Deoptimization of Foot-and-Mouth Disease Virus Causes Attenuation In Vivo while Inducing a Strong Neutralizing Antibody Response. Journal of virology. 2016;90:1298–1310. doi: 10.1128/JVI.02167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondili M, et al. Innate immune system activation by viral RNA: How to predict it? Virology. 2016;488:169–178. doi: 10.1016/j.virol.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Vabret N, et al. Large-scale nucleotide optimization of simian immunodeficiency virus reduces its capacity to stimulate type I interferon in vitro. Journal of virology. 2014;88:4161–4172. doi: 10.1128/JVI.03223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diebold SS, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 30.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 31.Forsbach A, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. Journal of immunology. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 32.Green NM, et al. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. The Journal of biological chemistry. 2012;287:39789–39799. doi: 10.1074/jbc.M112.383000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanji H, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nature structural & molecular biology. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Jurk M, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nature immunology. 2002;3:499. doi: 10.1038/ni0602-499. [DOI] [PubMed] [Google Scholar]
- 36.Ashman RF, et al. Optimal oligonucleotide sequences for TLR9 inhibitory activity in human cells: lack of correlation with TLR9 binding. International immunology. 2011;23:203–214. doi: 10.1093/intimm/dxq473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartmann G, Krieg AM. Mechanism and function of a newly identified CpG DNA motif in human primary B cells. Journal of immunology. 2000;164:944–953. doi: 10.4049/jimmunol.164.2.944. [DOI] [PubMed] [Google Scholar]
- 38.Ohto U, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 39.Rigby RE, et al. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. The EMBO journal. 2014;33:542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 41.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 43.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee MH, et al. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus replication. The Journal of general virology. 2011;92:2191–2200. doi: 10.1099/vir.0.032367-0. [DOI] [PubMed] [Google Scholar]
- 45.Saito T, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang C, et al. Sequence-Specific Modifications Enhance the Broad-Spectrum Antiviral Response Activated by RIG-I Agonists. Journal of virology. 2015;89:8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. Journal of virology. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Runge S, et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS pathogens. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez David RY, et al. Comparative analysis of viral RNA signatures on different RIG-I-like receptors. eLife. 2016:5. doi: 10.7554/eLife.11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baum A, et al. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu B, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 52.Yang Z, et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell research. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moy RH, et al. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158:764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herzner AM, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nature immunology. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia P, et al. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nature immunology. 2015;16:366–375. doi: 10.1038/ni.3117. [DOI] [PubMed] [Google Scholar]
- 59.Barreau C, et al. AU-rich elements and associated factors: are there unifying principles? Nucleic acids research. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wiley interdisciplinary reviews RNA. 2010;1:432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 61.Cathcart AL, et al. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. Journal of virology. 2013;87:10423–10434. doi: 10.1128/JVI.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S, et al. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nature structural & molecular biology. 2012;19:430–435. doi: 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- 64.Malathi K, et al. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y, et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dewannieux M, Heidmann T. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Current opinion in virology. 2013;3:646–656. doi: 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nature immunology. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature genetics. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5699–5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Q, et al. Type I interferon controls propagation of long interspersed element-1. The Journal of biological chemistry. 2015;290:10191–10199. doi: 10.1074/jbc.M114.612374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young GR, et al. Microarray analysis reveals global modulation of endogenous retroelement transcription by microbes. Retrovirology. 2014;11:59. doi: 10.1186/1742-4690-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hung T, et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng M, et al. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346:1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Karijolich J, et al. Infection-Induced Retrotransposon-Derived Noncoding RNAs Enhance Herpesviral Gene Expression via the NF-kappaB Pathway. PLoS pathogens. 2015;11:e1005260. doi: 10.1371/journal.ppat.1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu F, et al. Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nature reviews Cancer. 2016;16:359–372. doi: 10.1038/nrc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiappinelli KB, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roulois D, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanne A, et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bersani F, et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15148–15153. doi: 10.1073/pnas.1518008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song W, et al. Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13. Nature structural & molecular biology. 2015;22:782–787. doi: 10.1038/nsmb.3080. [DOI] [PubMed] [Google Scholar]
- 82.Li XD, Chen ZJ. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife. 2012;1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oldenburg M, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 84.Trotta E. On the normalization of the minimum free energy of RNAs by sequence length. PloS one. 2014;9:e113380. doi: 10.1371/journal.pone.0113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Hemert F, Berkhout B. Nucleotide composition of the Zika virus RNA genome and its codon usage. Virology journal. 2016;13:95. doi: 10.1186/s12985-016-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berkhout B, van Hemert F. On the biased nucleotide composition of the human coronavirus RNA genome. Virus research. 2015;202:41–47. doi: 10.1016/j.virusres.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alexopoulou L, et al. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 88.Leonard JN, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tatematsu M, et al. Beyond dsRNA: Toll-like receptor 3 signalling in RNA-induced immune responses. The Biochemical journal. 2014;458:195–201. doi: 10.1042/BJ20131492. [DOI] [PubMed] [Google Scholar]
- 91.Hull CM, Bevilacqua PC. Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR. Accounts of chemical research. 2016;49:1242–1249. doi: 10.1021/acs.accounts.6b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwok CK, et al. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends in biochemical sciences. 2015;40:221–232. doi: 10.1016/j.tibs.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Radhakrishnan A, et al. The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell. 2016;167:122–132. e129. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vabret N, Blander JM. Sensing microbial RNA in the cytosol. Frontiers in immunology. 2013;4:468. doi: 10.3389/fimmu.2013.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]