Fig. 1.
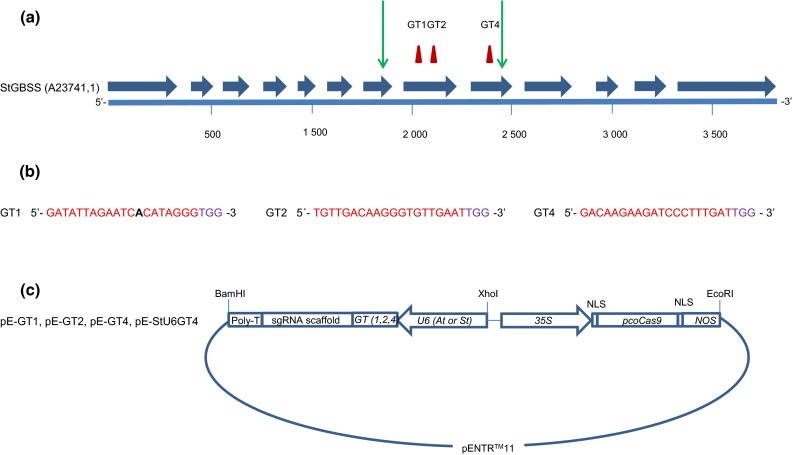
Design of CRISPR-Cas9 constructs targeting the GBSS gene. a Illustration of Solanum tuberosum GBSS gene structure (GenBank accession no. A23741.1). Exons 1–13 are marked with blue arrows, outer boundaries of fragments amplified for sequence determination of allelic variation are marked with green arrows and CRISPR-Cas9 target regions GT1, GT2 and GT4 are marked with red triangles. b Target regions GT1, GT2 and GT4 are in red and PAM-site in purple. Allelic variation in GT1 marked in black bold text. c Constructs designed for CRISPR-Cas9-mediated induction of mutations in StGBSS; from left to right: terminator (poly-T), sgRNA scaffold, guide sequence (GT1, GT2 or GT4), U6 promoter of either Arabidopsis thaliana or Solanum tuberosum origin, 35S promoter of cauliflower mosaic virus origin (CaMV), nuclear localization sequence (NLS), plant codon-optimized Cas9 gene, NLS and nopaline synthase terminator (NOS) in vector pENTR™11. The elements shown in the illustration are not to scale in relation to each other
