Fig. 3.
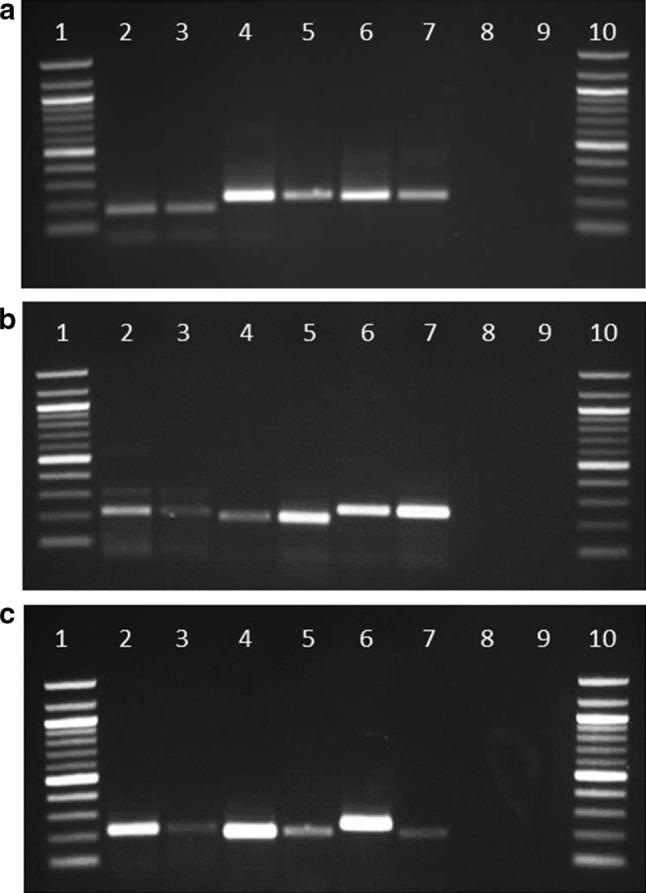
Reverse transcriptase-PCR assays of heat-, drought-, and salinity-responsive genes in switchgrass ecotypes, AP13 and VS16. Lane 1 and 10 100 bp ladder; Lanes 2, 4, 6 AP13 and 3, 5, 7 VS16; Lane 8 No reverse transcriptase (RT); Lane 9 H2O. a Heat stress-responsive genes: Heat shock protein 20 (Hsp20) (lanes 2–3), Heat shock protein 90 (Hsp90) (lanes 4–5), and Chaperone DnaJ-domain superfamily protein (lanes 6–7). b Drought stress-responsive genes: Delta tonoplast integral protein (2–3), Plasma membrane intrinsic protein 3 (4–5), and NOD-26 intrinsic protein 5;1 (6–7). c Salinity stress-responsive genes: Vacuolar H+-ATPase Subunit E isoform 3 (2–3), Leucine-rice repeat protein kinase family protein BRASSINOSTEROID INSENSITIVE 1 precursor (4–5), and Aldehyde dehydrogenase 2C4 (6–7). We used cons7 as an internal control for normalization. Note: Unusual posttranscriptional processing such as deletion of 5′ exon material or insertion of exogenous gene sequences was seen to result in differential transcriptional products in in cereal crops (Niu et al. 2007). We suggest that the distinct genetic differences between the upland VS16 genotype and lowland AP13 genotype used in this study could be a reason for differential product sizes of the aldehyde dehydrogenase gene
