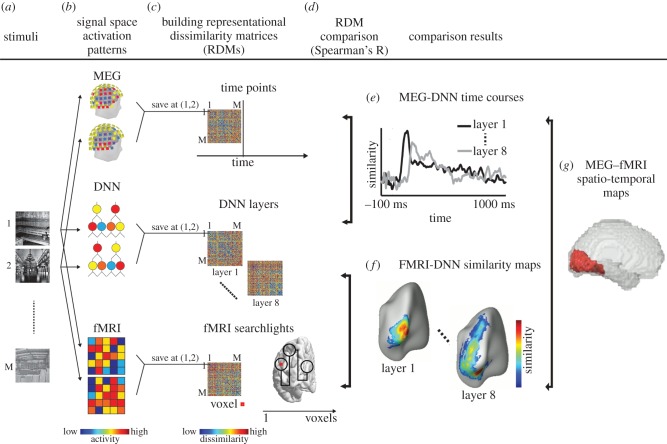
Figure 3.
RSA as a quantitative framework for combining models and data from different neuroimaging techniques. (a) A large conditions set (scene images) is used to probe brain activation in different neuroimaging techniques and also computational models (DNNs). (b) Condition-specific patterns are recorded for each signal space. (c) To make data directly comparable, signal-space patterns are transformed into dissimilarity space. For this, all pairwise combinations of conditions are compared with each other by dissimilarity (e.g. by calculating 1 minus Spearman's correlation, or decoding accuracy of a classifier), forming so-called representational dissimilarity matrices (RDMs), indexed in rows and columns by the conditions compared. RDMs are a summary of the representational geometry of the signal space, indicating which conditions evoke similar or different patterns. Such RDMs can be formed for each time point with technique with high temporal resolution such as M/EEG, for each location in cortical space with techniques that have high spatial resolution, such as fMRI, and for parts of computational models, such as layers of the DNNs. (d) In the similarity space of RDMs, fMRI, M/EEG and models can be linked to each other, combining their respective advantages, by calculating the similarity between RDMs (e.g. by calculating simple Spearman's correlation between RDMs). This yields (e) MEG–DNN time courses, (f) MEG–fMRI spatio-temporal maps and (g) fMRI–DNN similarity maps. Figure adapted from [35].