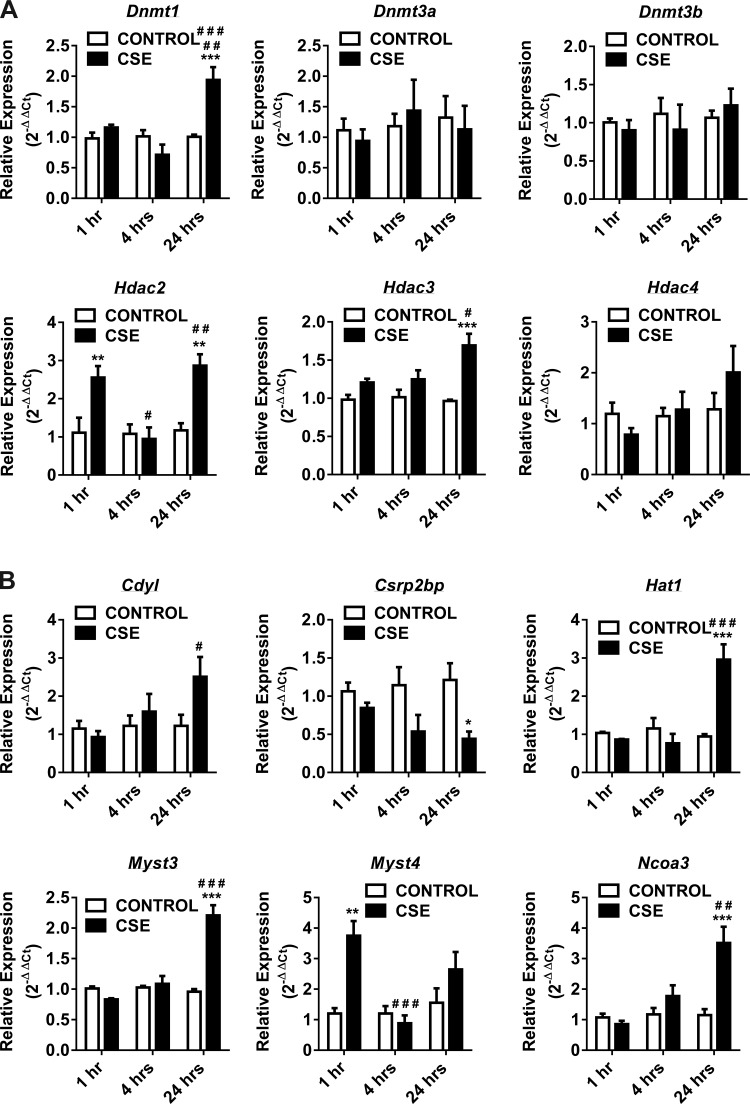
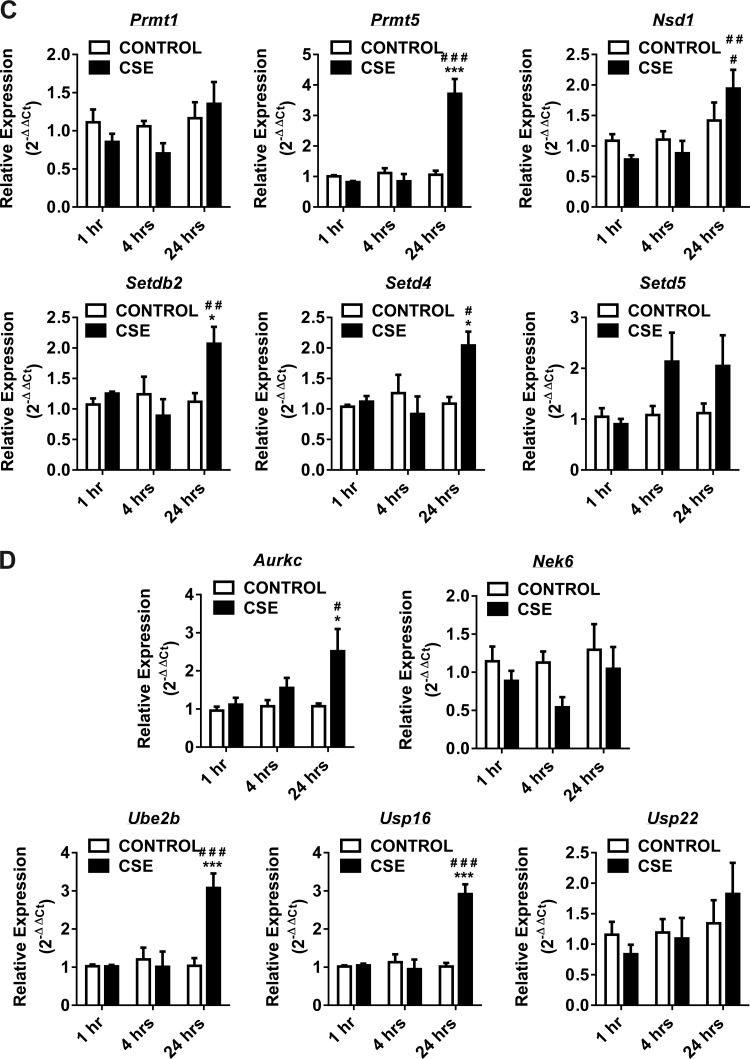
Fig. 2.
Gene expression of DNA methyltransferases, histone deacetylases, histone acetyltransferases, histone methyltransferases and SET domain proteins, histone phosphorylation, and histone ubiquitination enzymes at 1, 4, and 24 h in CSE-treated H292 cells. Human bronchial epithelial cells were treated with and without CSE for 1 h (2% CSE) and 4 and 24 h (1% CSE). RNA extracted from control and CSE-treated cells was analyzed using RT2 Profiler PCR array for human epigenetic chromatin modification enzymes. A: the transcription levels of genes encoding specific DNMTs (Dnmt1, Dnmt3a, and Dnmt3b) and HDACs (Hdac2, Hdac3, and Hdac4) were examined by qPCR using the 2−ΔΔCt method. B: the transcription levels of genes encoding specific HATs (Cdyl, Csrp2bp, Hat1, Myst3, Myst4, and Ncoa3) were examined by qPCR using the 2−ΔΔCt method. C: the transcription levels of genes encoding specific HMTs (Prmt1, Prmt5, and Nsd1) and SET domain proteins (Setdb2, Setd4, and Setd5) were examined by qPCR using the 2−ΔΔCt method. D: the transcription levels of genes encoding specific histone phosphorylation (Aurkc and Nek6) and histone ubiquitination (Ube2b, Usp16, and Usp22) were examined by qPCR using the 2−ΔΔCt method. Bar graphs represent the mean normalized expression of samples in control vs. CSE-treated H292 cells. Data were normalized using the endogenous housekeeping gene ribosomal protein L13a (Rpl13a) as reference and controls as calibrators. Statistical significance (P < 0.05) was analyzed by two-way ANOVA (Tukey's multiple-comparison test) using GraphPad Prism 6. Values are means ± SE (n = 4/group). *P < 0.05, **P < 0.01, and ***P < 0.001 vs. control. #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. CSE (1 or 4 h).