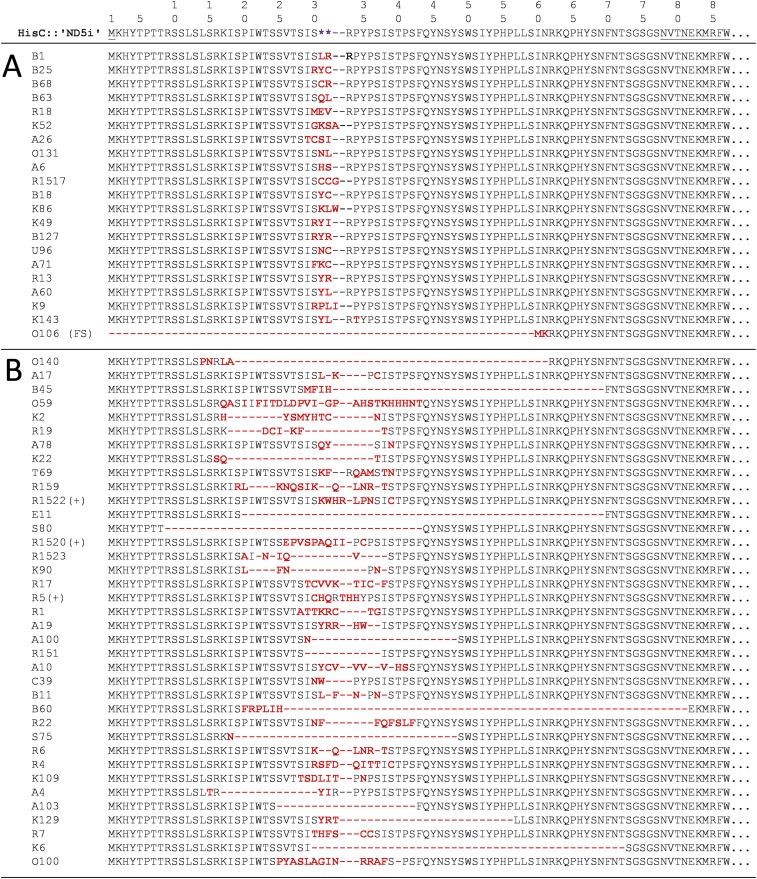
Fig. S2.
Deduced amino acid sequence alignments from the ′ND5i′ insert of the recombinant HisC from all experimentally obtained A. baylyi HisC+ revertant SPDIR isolates. The stop codons in the hisC::′ND5i′ detection allele are depicted as purple asterisks, and codon changes are indicated in red. The wt HisC codons flanking the ′ND5i′ insert are underlined in the top line. All corresponding DNA sequences are given in Dataset S1. (A) Class 1 SPDIR events formed at a single, contiguous microhomology typically contain two to four variable codon changes without change in protein length. In isolate O106 (Fig. 1C), the replacement introduced a frameshift (FS). (B) Illegitimate joints occurring at separate simple or extended microhomologies (class 2 SPDIR events) resulted in highly variable changes often associated with deletions. Events with total net amino acid gains are marked with (+).