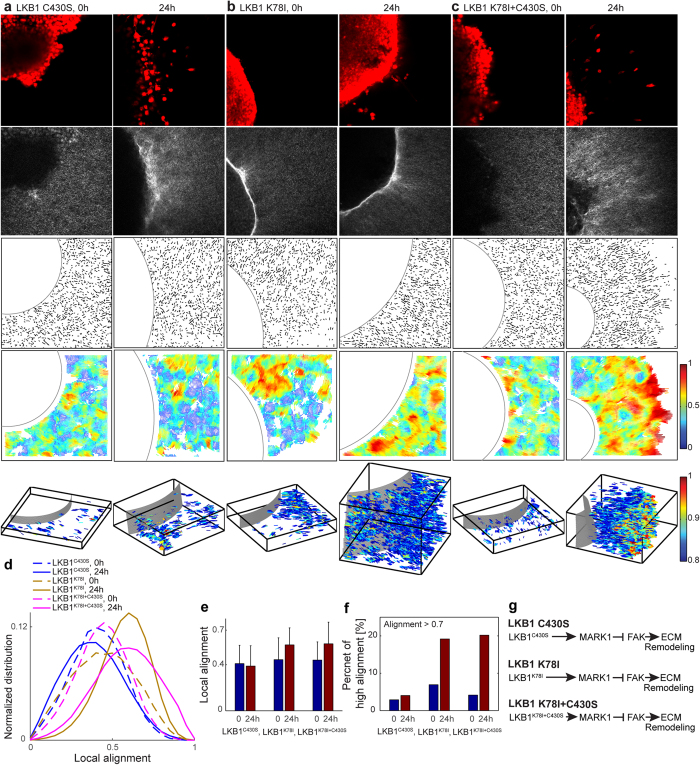
Figure 6. Local alignment vector analysis of in vitro H157 tumor spheroids in a collagen gel.
(a) LKB1C430S (farnesylation mutant), (b) LKB1K78I (kinase-dead mutant), (c) LKB1K78I + C430S (double mutant in kinase domain and farnesylation domain) cases at 0 and 24 hour after seeding a tumor spheroid into a 2 mg/ml collagen gel. The first row in (a–c) is multiphoton microscopy images to visualize tumor spheroids, dyed using CellTracker Red; the second row is multiphoton microscopy images to visualize collage fibers using second harmonic generation signals; the third row is quantized fiber segments with manually selected circular tumor boundary after extracting collagen fibers using CT-FIRE; the fourth row is local alignment vectors. These 2D fiber images are the first z stack images (the bottom of collagen gel). The fifth row is local alignment vectors of all z stacks in 3D, where we plt only high alignment vectors (alignment > 0.8). For the clear visualization, we magnify 4 times in Z, e.g. X:Y:Z = 1:1:4. (d) Normalized local alignment distribution using all z stack data. The local alignment (e) and the high alignment ratio (f) over the whole z stacks were calculated. (g) LKB1 signaling pathway for LKB1C430S, LKB1K78I, and LKB1K78I + C430S case.