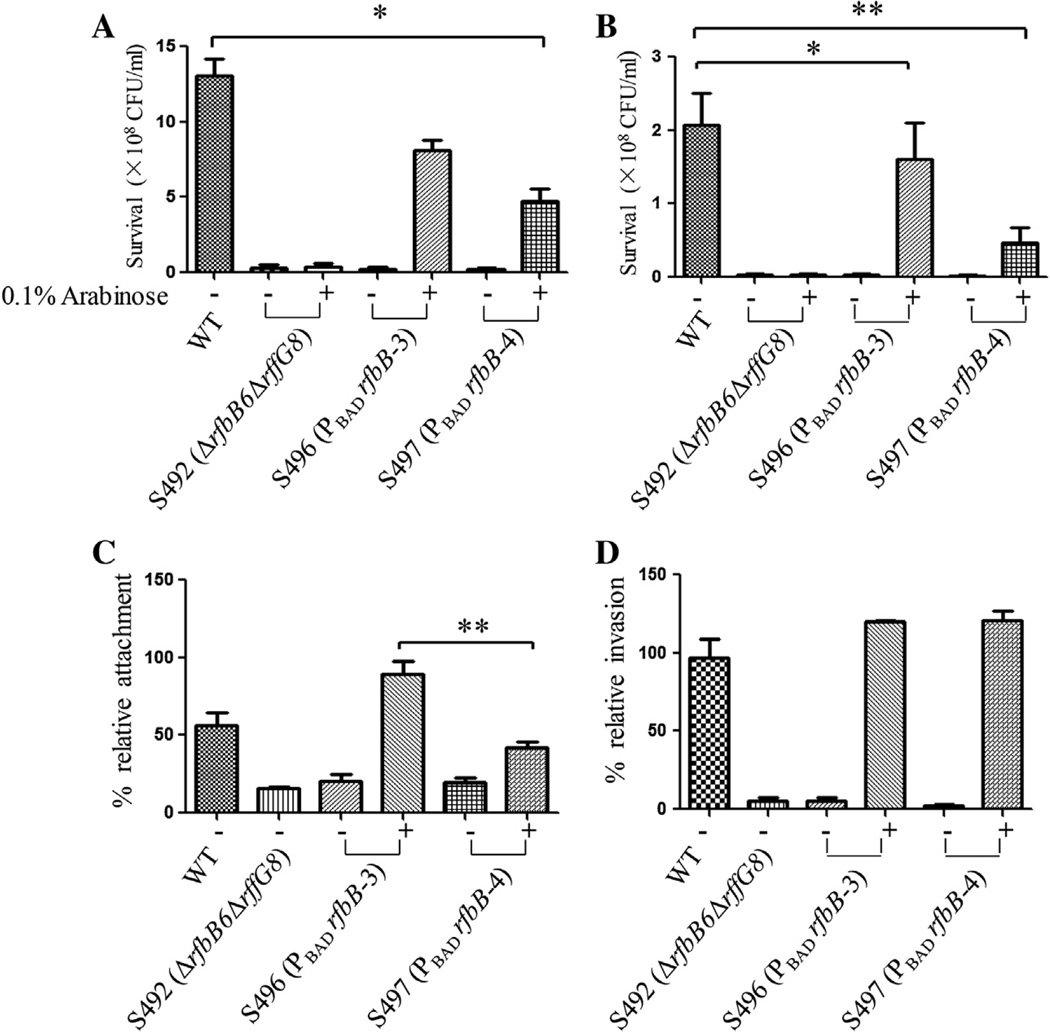
Fig. 2.
Polymyxin B and DOC sensitivity and attachment/invasion assays for the mutant strains. (A) Polymyxin B sensitivity assay. Approximately 1 × 107 CFU/ml bacteria with 0.1 µg/ml polymyxin B were incubated for 60 min at 37 °C. The bacterial survival ratio was calculated after plating the bacteria on LB agar and counting colonies the following day. Values that are significantly different from those of the wild type as indicated by one-way ANOVA are indicated by asterisks (*P < 0.05). (B) DOC sensitivity assay. Approximately 1 × 107 CFU/ml bacteria with 10 mg/ml DOC were incubated for 60 min at 37 °C. The bacterial survival ratio was calculated after plating the bacteria on LB agar and counting colonies the following day. Values that are significantly different from those of the wild type as indicated by one-way ANOVA are indicated by asterisks (*P < 0.05, **P < 0.01). (C) Attachment efficiency of the Salmonella mutants to INT407 cells. The mutant strains were added to wells with INT407 cells to achieve an MOI of 5:1. A 1% Triton X-100 solution was added to lyse the INT407 cells after incubation for 60 min at 37 °C. After plating on petri dishes and overnight incubation, the colony counts were determined. Attachment was calculated as follows: percent attachment = 100 × (number of cell-associated bacteria/initial number of bacteria added). Values that are significantly different as determined by one-way ANOVA are indicated by asterisks (**P < 0.01). (D) Invasion efficiency of Salmonella mutants to INT407 cells. Prior to the lysis step, 100 µg/ml gentamicin was added to kill the extracellular bacteria, and the plate was incubated for an additional 60 min at 37 °C. Invasion was calculated as follows: percent invasion = 100 × (number of bacteria resistant to gentamicin/initial number of bacteria added).