Abstract
Background & objectives:
Non-fermenting Gram-negative bacilli (NFGNB) including Pseudomonas aeruginosa and Acinetobacter baumannii have been implicated in a variety of infections, particularly in the Intensive Care Units (ICUs). This study was aimed to overview the burden of multidrug-resistant NFGNB causing infections in ICU and also to assess the occurrence of extended-spectrum beta-lactamases (ESBLs), AmpC and metallo-beta-lactamases (MBLs) among these isolates.
Methods:
Bacterial culture, identification and antibiotic susceptibility were carried out. ESBLs and AmpC were detected both phenotypically and genotypically. MBL was detected by modified Hodge and imipenem-ethylenediaminetetraacetic acid double-disc synergy test.
Results:
NFGNB represented 45 (37%) of total 121 Gram negative isolates. Multidrug resistance was observed in 66.9 per cent and 72.5 per cent isolates of P. aeruginosa and A. baumannii, respectively. Detection by phenotypic methods showed presence of ESBL, AmpC and MBL in 21.4, 51.1 and 21.4 per cent isolates, respectively. When detected genotypically by polymerase chain reaction, ESBL and AmpC were detected in 21.4 and 41.4 per cent of NFGNB isolates, respectively. BlaCTX-M (21.4%) was the most prevalent gene responsible for ESBL production.
Interpretation & conclusions:
Most of the NFGNB isolated from ICU patients were multidrug-resistant and producers of ESBL, AmpC and MBL. A regular surveillance is required to detect ESBL, AmpC and MBL producers, especially in ICU patients.
Keywords: Acinetobacter baumannii, Intensive Care Unit, multidrug-resistance, Pseudomonas aeruginosa
Non-fermenting Gram-negative bacilli (NFGNB) including Pseudomonas aeruginosa and Acinetobacter baumannii have been implicated in a variety of infections, including bacteraemia, urinary tract and surgical site infections among patients admitted in Intensive Care Unit (ICU)1,2. These may be intrinsically resistant or may have acquired resistance to antibiotics due to impermeability of the cell surface, multidrug efflux pumps and production of β-lactamases [AmpC β-lactamase, extended-spectrum β-lactamases (ESBLs) and metallo-beta-lactamases (MBLs)]3. Multiple beta-lactamase-producing P. aeruginosa can cause major therapeutic failure and poses a significant clinical challenge. Reports on carbapenemase-producing NFGNB are on the rise globally due to the increased carbapenem usage and selection of resistant bacteria under antibiotic pressure4. Therefore, early identification and detection of isolates that produce these enzymes are essential to avoid therapeutic failures and nosocomial outbreaks.
This study was designed to assess the burden of multidrug-resistant P. aeruginosa and A. baumannii in ICU patients. The occurrence of ESBL, AmpC and MBL among these isolates was also assessed.
Material & Methods
The present study was carried out in the department of Microbiology on patients admitted to the ICU of J. N. Medical College, Aligarh Muslim University, Aligarh, India from February 2012 to October 2013. Totally, 125 patients admitted to the ICU were included in the study. A complete history was taken from each patient. Informed written consent was obtained before the study from all the patients, and the study was performed after getting approval from the Institutional Ethics Committee.
The patients were chosen consecutively, and clinical samples were obtained from each patient (endotracheal aspirate, blood, pus, urine). All specimens were collected aseptically and were promptly sent to the microbiology laboratory. All samples were collected within 48 h of the patient admission in the ICU and those collected after 48 h of admission were not included in the study. Wounds (surgical site infections) have been classified according to the Southampton grading5. The majority of the cases belonged to Grade IV (purulent discharge along the wound) and Grade V (wound dehiscence). Standard methods for isolation and identification of NFGNB6 were used.
Susceptibility testing of bacterial isolates was performed using the disc diffusion method as described by the Clinical and Laboratory Standards Institute7. Antimicrobial discs used were imipenem (10 µg), cefpodoxime (10 µg), cefotaxime (30 µg), cefepime (30 µg), cefixime (5 µg), cefoperazone (75 µg), cefoperazone/sulbactam (75/10 µg), ticarcillin (75 µg), piperacillin (100 µg), piperacillin/tazobactam (100/10 µg), ceftazidime (30 µg), ceftazidime/clavulanic acid (30/10 µg), cefotaxime/clavulanic acid (30/10 µg), ceftriaxone (30 µg), amikacin (30 µg), gentamicin (10 µg), tobramycin (10 µg), ofloxacin (5 µg), levofloxacin (5 µg), polymixin B (300 units) and colistin (10 µg). All discs were obtained from Hi-Media Labs, Mumbai, India.
Phenotypic methods for ESBL detection: NFGNB isolates were first screened for the production of ESBL by the disc diffusion method (screening test) using cefotaxime, ceftriaxone, cefepime and ceftazidime7 and later on confirmed by the cephalosporin/clavulanate combination disc (disc potentiation test)8 and double-disc synergy test8,9. Escherichia coli ATCC 25922 (non-ESBL producer) was used as a control strain.
Phenotypic methods for AmpC detection: Cefoxitin discs were used to screen AmpC producers, by disc diffusion method10. Isolates resistant to cefoxitin were considered as potential AmpC producers.
Phenotypic methods for MBL detection: The isolates were tested for sensitivity to imipenem (10 μg) using Kirby-Bauer method as recommended by the CLSI7. All the isolates with a zone of inhibition ≤16 mm or which demonstrated heaping, or if the zone was >16 but ≤20 mm, were tested for MBL production; however, there is no CLSI guideline for MBL detection available for P. aeruginosa. These isolates were confirmed by modified Hodge test and imipenem-ethylenediaminetetraacetic acid (EDTA) double-disc synergy test11,12.
Genotypic methods for the detection of ESBL and AmpC production: Template DNA was prepared from freshly cultured bacterial isolates by suspending bacterial colonies in 50 µl of molecular grade water and then heating at 95°C for five minutes and immediately chilling at 4°C. Molecular detection of blaCTX-M, blaTEM, blaSHV and blaAmpC was performed using polymerase chain reaction (PCR) according to methods described previously with minor modifications (thermal profile of blaCTX-M12, primer profiles of blaCTX-M, blaTEM, blaSHV, blaAmpC13). The primers and cycling conditions for detection of blaAmpC genes were the same as those described by Shahid et al12 and Féria et al13. The quality-control strain, Klebsiella pneumoniae ATCC 700603 (ESBL producer) was used.
Results & Discussion
Among the 125 patients admitted to the ICU, 160 isolates were identified. Of these, Gram-negative bacilli, 121 (75.6%) predominated, followed by 22 (13.8%) Gram-positive cocci and 10.6 per cent (n=17) fungal isolates. NFGNB represented 45 (37%) of the Gram-negative isolates (n=121) of which P. aeruginosa (n=35, 29%) was the incriminatory pathogen in majority, followed by A. baumannii (n=10, 8%).
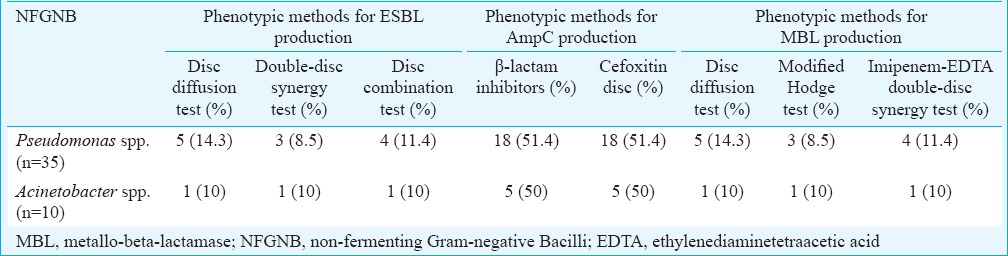
Antimicrobial resistance was observed to be higher in A. baumannii than in P. aeruginosa. Antibiotic resistance pattern of P. aeruginosa and A. baumannii is shown in Table I. Table II depicts the organisms isolated from different samples of patients in ICU. The positivity for ESBL, AmpC and MBL by phenotypic methods is shown in Table III.
Table I.
Antibiotic resistance pattern of 45 non-fermenting Gram-negative bacilli detected by disc diffusion method
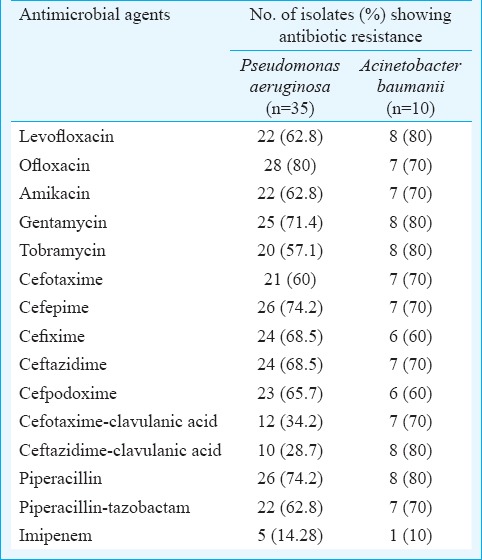
Table II.
Distribution of pathogens isolated from endotracheal aspirate and urinary tract infection in Intensive Care Unit patients
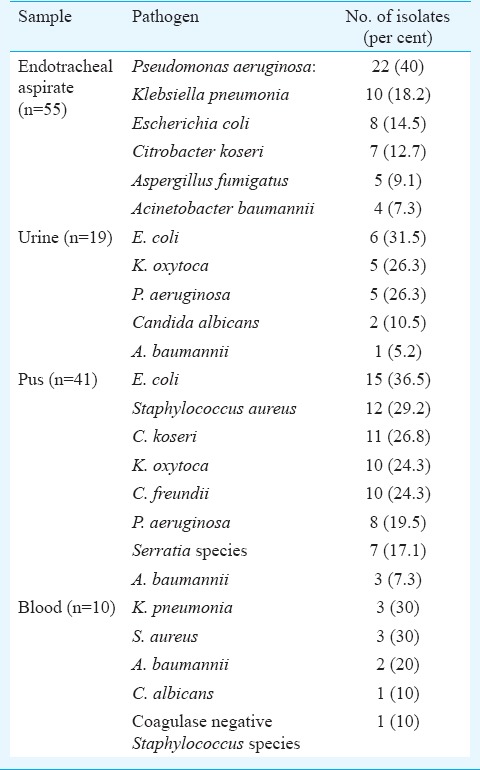
Table III.
Number and percentage of extended-spectrum β-lactamase (ESBL)-producing Pseudomonas aeruginosa and Acinetobacter baumannii by phenotypic methods
BlaCTX-M, blaSHV, blaTEM and blaAmpC genes were detected in phenotypic ESBL and AmpC producers. BlaCTX-M was the predominant gene among ESBL producers as it was observed in four (11.4%) isolates of P. aeruginosa and one (10%) A. baumannii. BlaAmpC was detected in 15 (42.8%) P. aeruginosa and four (40%) A. baumannii isolates. BlaSHV and blaTEM were not detected in any of the isolates.
NFGNB including P. aeruginosa and A. baumannii have been implicated in a variety of ICU infections. In this study, P. aeruginosa represented 29 per cent of isolates; similar results were reported by Hadadi et al14. In other studies, P. aeruginosa represented 15.6 per cent of the total isolates15,16. A. baumannii represented eight per cent of Gram-negative ICU infections in this study. However, other investigators found a higher incidence of A. baumannii (20.5 and 24.1%, respectively)14,15.
In this study, lower respiratory tract infections (LRTIs) were the most common infection in ICU patients (34.3%). Al-Ghamdi et al17 reported 8.9 per cent LRTIs among ICU cases. In our study, P. aeruginosa and A. baumannii were frequently isolated from LRTIs (40 and 7.3%, respectively), as also shown by Abd El-Fattah18.
In our study 76.1 and 70.2 per cent of NFGNB were resistant to fluoroquinolones and aminoglycosides, respectively. Antibiotic resistance is a serious problem in developing countries, especially due to the easy availability of antibiotics over the counter19,20. The resistance of P. aeruginosa and A. baumannii to ceftazidime (68.5%, 70%), cefotaxime (57.1%, 70%) and cefpodoxime (65.7%, 70%) was comparable to results reported by others14,21.
Screening by disc diffusion method in this study revealed that 24.3 per cent of NFGNB were ESBL producers. These results were comparable to that of Aggarwal et al22, however, lower than results reported by Jiang et al23. Confirmation with double-disc synergy test (DDST) and disc combination tests revealed that 18.5 per cent of NF Gram-negative isolates were ESBL producers as also reported by Jiang et al23. For AmpC production, both disc diffusion test and screening by cefoxitin disc revealed same results that 51.4 per cent of P. aeruginosa and 50 per cent of A. baumannii isolates were AmpC producers. Similar results were reported by Bhattacharjee et al24. The most prevalent gene for ESBL production was blaCTX-M which was detected in 11.4 per cent of Pseudomonas isolates, while blaTEM and blaSHV genes were not detected in any of the isolates. These results were in agreement with Picão and Gales25. PCR detected blaAmpC gene in 42.8 per cent of P. aeruginosa and 40 per cent of A. baumannii isolates. These results were comparable to that of Khanal et al26.
Comparison between modified Hodge test and DDST in our study revealed that DDST was more sensitive for detecting MBL. The same observation was reported by Jesudason et al27. In this study, amikacin, tobramycin, imipenem, polymyxin B and colistin demonstrated maximum sensitivity against NFGNB. Therefore, use of these antibiotics should be restricted to severe infections, especially in critically ill ICU patients, to avoid rapid emergence of resistant strains.
In conclusion, our results showed isolation of a high percentage of NFGNB in ICU patients’ samples, which were multidrug resistant and producers of ESBL, AmpC and MBL. A regular surveillance of antimicrobial susceptibility status of such isolates is necessary to curb the infection.
References
- 1.Nseir S, Di Pompeo C, Brisson H, Dewavrin F, Tissier S, Diarra M, et al. Intensive care unit-acquired Stenotrophomonas maltophilia: incidence, risk factors, and outcome. Crit Care. 2006;10:R143. doi: 10.1186/cc5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malini A, Deepa E, Gokul BN. Nonfermenting Gram-negative bacilli infections in a tertiary care hospital in Kolar, Karnataka. J Lab Physicians. 2009;1:62–6. doi: 10.4103/0974-2727.59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50:43–8. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gales AC, Jones RN, Forward KR, Liñares J, Sader HS, Verhoef J. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997-1999) Clin Infect Dis. 2001;32 (Suppl 2):S104–13. doi: 10.1086/320183. [DOI] [PubMed] [Google Scholar]
- 5.Wilson APR, Gibbons C, Reeves BC, Hodgson B, Liu M, Plummer D. Surgical wound infection as a performance indicator: agreement of common definitions of wound infection in 4773 patients. BMJ. 2004;329:720. doi: 10.1136/bmj.38232.646227.DE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collee JG, Marr W. Culture of bacteria. In: Fraser AG, Collee JG, Marmion BP, Simmons A, editors. Mackie and McCartney practical microbiology. 14th ed. London: Churchill Livingstone; 1996. pp. 113–30. [Google Scholar]
- 7.Performance standards for antimicrobial susceptibility testing: Seventeenth informational supplement. M100-S17. Wayne, PA: CLSI; 2007. Clinical and Laboratory Standards Institute (CLSI) [Google Scholar]
- 8.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–78. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. bla(VIM-2) cassette-containing novel integrons in metallo-beta-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 2002;46:1053–8. doi: 10.1128/AAC.46.4.1053-1058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahid M, Malik A, Agrawal M, Singhal S. Phenotypic detection of extended-spectrum and AmpC beta-lactamases by a new spot-inoculation method and modified three-dimensional extract test: comparison with the conventional three-dimensional extract test. J Antimicrob Chemother. 2004;54:684–7. doi: 10.1093/jac/dkh389. [DOI] [PubMed] [Google Scholar]
- 12.Shahid M, Ensor VM, Hawkey PM. Emergence and dissemination of Enterobacteriaceae with plasmid-mediated CMY-6 and CTX-M-15 beta-lactamases in a community in North-India. World J Microbiol Biotechnol. 2009;25:1439–46. [Google Scholar]
- 13.Féria C, Ferreira E, Correia JD, Gonçalves J, Caniça M. Patterns and mechanisms of resistance to beta-lactams and beta-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. 2002;49:77–85. doi: 10.1093/jac/49.1.77. [DOI] [PubMed] [Google Scholar]
- 14.Hadadi A, Rasoulinejad M, Maleki Z, Yonesian M, Shirani A, Kourorian Z. Antimicrobial resistance pattern of gram-negative bacilli of nosocomial origin at 2 university hospitals in Iran. Diagn Microbiol Infect Dis. 2008;60:301–5. doi: 10.1016/j.diagmicrobio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Chim H, Tan BH, Song C. Five-year review of infections in a burn intensive care unit: high incidence of Acinetobacter baumannii in a tropical climate. Burns. 2007;33:1008–14. doi: 10.1016/j.burns.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Izquierdo-Cubas F, Zambrano A, Frómeta I, Gutiérrez A, Bastanzuri M, Guanche H, et al. National prevalence of nosocomial infections. Cuba 2004. J Hosp Infect. 2008;68:234–40. doi: 10.1016/j.jhin.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Al-Ghamdi S, Gedebou M, Bilal NE. Nosocomial infections and misuse of antibiotics in a provincial community hospital, Saudi Arabia. J Hosp Infect. 2002;50:115–21. doi: 10.1053/jhin.2001.1149. [DOI] [PubMed] [Google Scholar]
- 18.Abd El-Fattah SM. Faculty of Medicine Giza. Egypt: Cairo University; 2008. Evaluation of antibiotic resistance among Gram-ve bacilli isolated from critically ill patients: Relation to risk factors and liberal use of antibiotics. M.Sc Thesis in Medical Microbiology and Immunology. [Google Scholar]
- 19.Giamarellou H, Antoniadou A, Kanellakopoulou K. Acinetobacter baumannii: a universal threat to public health? J Hosp Infect. 2007;67:245–52. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Martínez JL, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15:647–79. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGowan JE., Jr Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am J Med. 2006;119(Suppl 1):S29–36. doi: 10.1016/j.amjmed.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal R, Chaudhary U, Bala K. Detection of extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Indian J Pathol Microbiol. 2008;51:222–4. doi: 10.4103/0377-4929.41693. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Zhang Z, Li M, Zhou D, Ruan F, Lu Y. Detection of extended-spectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:2990–5. doi: 10.1128/AAC.01511-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharjee A, Anupurba S, Guar A, Sen MR. The prevalence of inducible AmpC β-lactamase producing Pseudomonas aeruginosa in a tertiary care hospital in northern India. Indian J Med Microbial. 2008;26:89–98. doi: 10.4103/0255-0857.38872. [DOI] [PubMed] [Google Scholar]
- 25.Picão RC, Gales AC. Extended-spectrum beta-lactamase-producing (ESBL) Pseudomonas aeruginosa: nightmare or imagination? Prát Hosp. 2007;49:79–84. [Google Scholar]
- 26.Khanal S, Joshi DR, Bhatta DR, Devkota U, Pokhrel BM. β-lactamase-producing multidrug-resistant bacterial pathogens from tracheal aspirates of intensive care unit patients at national institute of neurological and allied sciences, Nepal. ISRN Microbiol. 2013;2013:847569. doi: 10.1155/2013/847569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jesudason MV, Kandathil AJ, Balaji V. Comparison of two methods to detect carbapenemase & metallo-beta- lactamase production in clinical isolates. Indian J Med Res. 2005;121:780–3. [PubMed] [Google Scholar]


