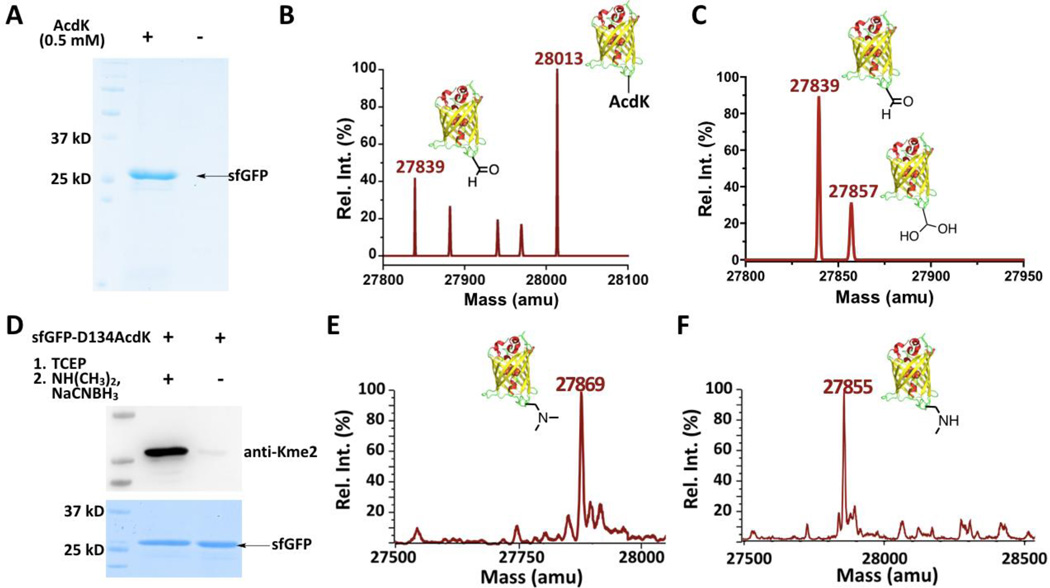
Figure 2. The genetic incorporation of AcdK into a model protein sfGFP for the lysine di- and monomethylation installation.
(A) The site-specific incorporation of AcdK into sfGFP at its D134 position to produce sfGFP-D134AcdK. Cells were transformed with two plasmids containing genes for AcdKRS, tRNAPyl, and sfGFP with an amber mutation at its D134 position and grown in LB media supplemented with or without 0.5 mM AcdK. (B–C) The ESI-MS spectra of purified sfGFP-S2AcdK (B) and its TCEP-reduced product (C). (D) The selective installation of Kme2 at the D134 position of sfGFP. SfGEP-D134AcdK was reduced with 5 mM TCEP in PBS buffer at pH 7 for 2 h followed by the reaction with 100 mM dimethylamine and 5 mM NaCNBH3 in PBS buffer at pH 7 for 8 h for the installation of Kme2 to form sfGEP-D134Kme2. The original sfGEP-D134AcdK was used as a control in the Western blot and SDS-PAGE analysis. The top panel shows a western blot probed by anti-Kme2 and the bottom panel shows a Coomassie blue stained gel for two proteins. (E–F) The ESI-MS spectraof sfGFP-D134Kme2 (E) and sfGFP-D134Kme (F).