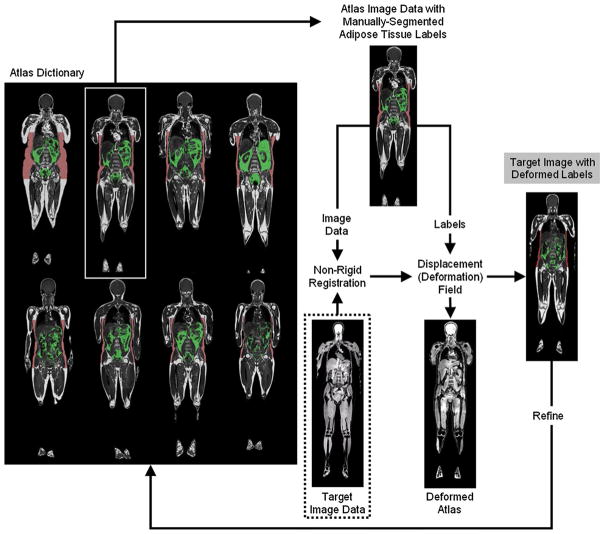
Fig. 5.
A schematic of atlas-based automated segmentation subcutaneous (red) and visceral (green) of adipose tissues. The target data set to be automatically segmented (dotted box) is registered to pre-existing volumes in the atlas, which represents a collection of manually segmented “gold-standard” reference data sets. The non-rigid registration step yields a displacement map, which is then applied to the atlas labels. Multiple atlases can be used to refine the procedure iteratively. In this particular example, the data sets were acquired with chemical-shift encoded water–fat MRI and all data sets have been pre-processed to correct for signal intensity bias field. Note that in this particular case, the water-only image from the target is fed into the non-rigid registration step. Illustration courtesy of Anette Karlsson and Olof Dahlqvist Leinhard, Linköping University (see Ref. [165] for details)