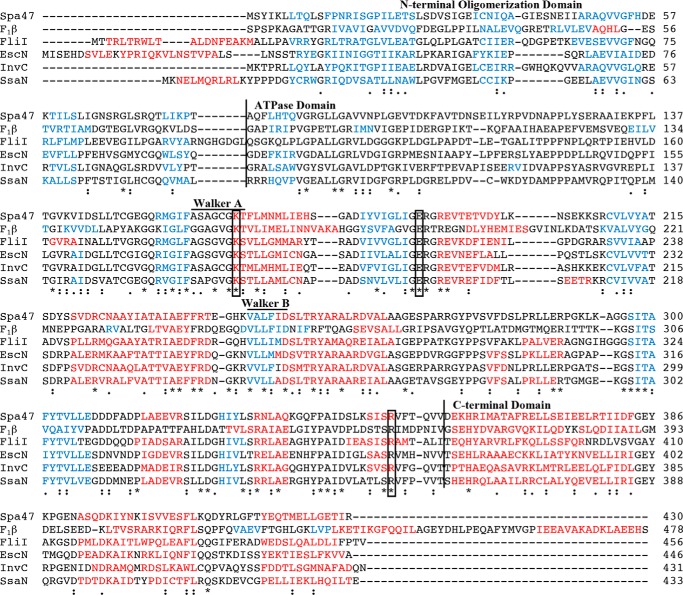
FIGURE 1.
Multiple sequence alignment of Spa47 and related ATPase homologs. Protein sequence alignment of Spa47 (T3SS ATPase, S. flexneri), F1β subunit (ATP synthase, Bos taurus), FliI (Flagellar ATPase, Salmonella typhimurium), EscN (T3SS ATPase, E. coli), InvC (T3SS ATPase, Salmonella gallinrium), and SsaN (T3SS ATPase, S. typhimurium) was performed using the Uniprot multiple sequence alignment tool, Clustal Omega, with single fully conserved residues (*), conservation between groups with strongly similar properties (:), and weakly similar properties (.) identified. Helical and β-sheet regions, as predicted by the PSIPRED structure prediction server, are color-coded red and blue, respectively. Vertical lines separate the predicted N-terminal oligomerization, ATPase, and C-terminal domains. Predicted Walker A (P-loop) and Walker B regions and the fully conserved residues targeted for alanine screening in this study are also identified in the alignment. Uniprot accession numbers used for sequence alignment were Q6XVW8, P00829, P26465, Q7DB71, B5RDL8, P74857 for Spa47, F1 ATPase B subunit, FliI, EscN, InvC, and SsaN, respectively.