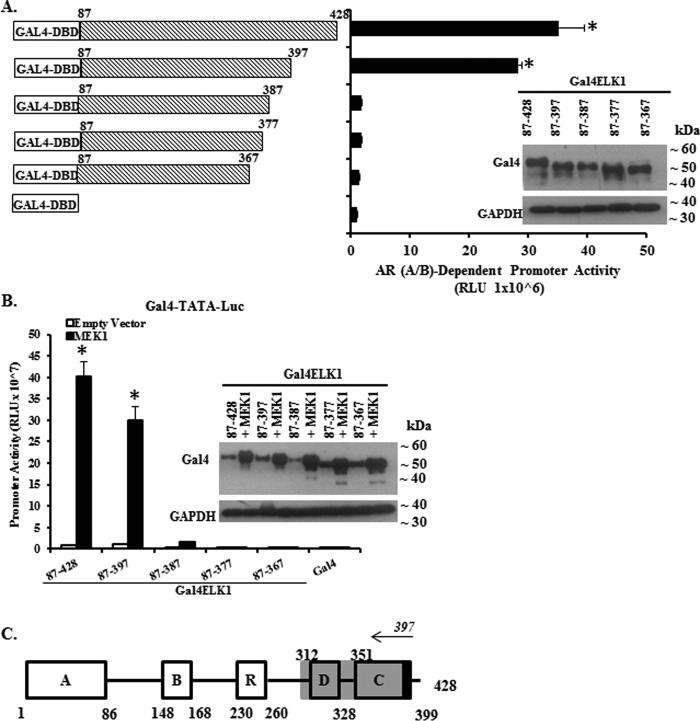
FIGURE 3.
Mapping ELK1 polypeptide segments required for co-activation by AR(A/B) by carboxyl-terminal deletion analysis. A shows data obtained using recombinant HeLa cells generated by stably transducing a minimal promoter-luciferase reporter containing upstream Gal4 elements (GAL4-TATA-LUC) and also with a vector expressing the AR A/B domain fused to the VP16 transactivation domain. Cells were transfected with plasmids expressing Gal4 fusion proteins of ELK1. The fusion constructs substituted the Gal4 DNA binding domain (Gal4-DBD) for the ETS DNA binding domain of ELK1. Within this fusion construct, a series of carboxyl-terminal deletions were made, as indicated in the schematic in A. Forty eight hours after transfection with the various Gal4-ELK1 fusion constructs, cells were harvested by preparing lysates for measurement of luciferase activity. The promoter activity shown on the y axis required the presence of the AR A/B domain because knocking down AR(A/B) expression in the same cells transfected with full-length Gal4-ELK1 decreased the promoter activity to the basal value shown in the figure for Gal4-DBD alone. The inset shows cell lysates probed by Western blotting with antibodies against Gal4 or GAPDH (loading control). B shows data obtained using recombinant HeLa cells generated by stably transducing only GAL4-TATA-LUC. The cells were transfected with each of the Gal4-ELK1 fusion constructs used in A and co-transfected with an expression plasmid for a constitutively active mutant of MEK1 or with the vector control. Forty eight hours after transfection with the various Gal4-ELK1 fusion constructs, cells were harvested by preparing lysates for measurement of luciferase activity. The inset shows cell lysates probed by Western blotting with antibodies against Gal4 or GAPDH (loading control). C shows a schematic of the domain organization of ELK1; here, the carboxyl-terminal deletion mapping of an ELK1 polypeptide segment encompassing residues required for association with AR(A/B) (data from A) is represented by gray shading. For all transfections, a Renilla luciferase reporter was used as the control for transfection efficiency. In all panels, the error bars represent standard deviation of experimental triplicates. *, p < 0.001.