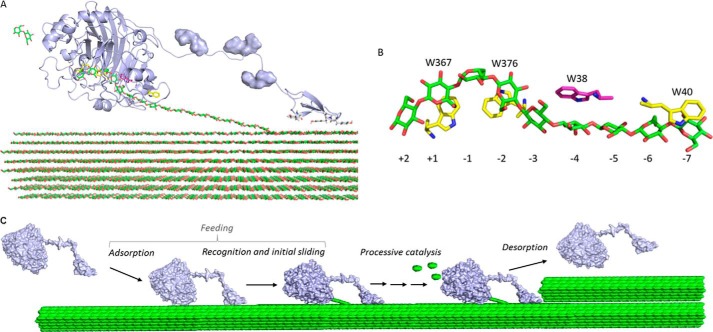
FIGURE 1.
Structure and mechanism of TrCel7A. A, schematic representation (light blue) of catalytic domain, linker, and CBM for TrCel7A in complex with a cellulose strand. The O-glycosylation of the linker is shown as a surface representation and the cellulose strand is represented as green sticks. The image was made using the crystal structure of the catalytic domain (Protein Data Bank code 8CEL) and CBM (Protein Data Bank code 1CBH). B, positions of the Trp residues in the active site tunnel of TrCel7A in complex with a cellodextrin chain (green). Numbers refer to the different binding subsites in the tunnel with −7 at the entrance and −1/+1 being the position of the scissile bond. Trp-38 investigated in this study is highlighted in magenta. C, molecular steps in the hydrolysis of cellulose for a processive enzyme. Steps leading from the free enzyme in the solution to the enzyme with the reducing end of the cellulose chain in the −1 binding site are collectively referred to as feeding. Processive catalysis includes the formation of a Michaelis complex (by sliding the chain end from binding site −1 to +2), hydrolysis of glycosidic bond, and expulsion of cellobiose (green ellipses). Processive catalysis is repeated until the enzyme meets an obstacle (depicted here as upper cellulose fibril) or happens to dissociate.