FIGURE 6.
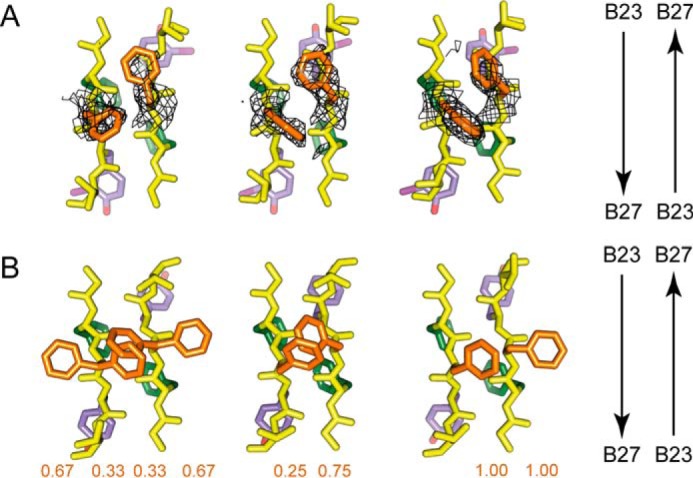
Side-chain arrangements within dimer interfaces. Residues B23–B26 are shown within respective crystal structures of the 3-[iodo-TyrB26,NleB29]insulin hexamer (A) and WT insulin R6 zinc hexamer (PDB code 1ZNJ) (B). The three subpanels within A and B correspond to the respective three copies of the dimer interface within the crystallographic asymmetric units of the two structures. Within each subpanel, the side-chain carbon atoms of PheB24 and its non-crystallographic symmetry equivalents are shown in green, of PheB25 and its non-crystallographic symmetry equivalents in orange, and of 3-I-TyrB26 or TyrB26 and their non-crystallographic symmetry equivalents in light purple, whereas all backbone atoms are in yellow, as are the side-chain atoms of ThrB27 and its non-crystallographic symmetry equivalents. The arrows on the right assist in identifying the direction of the respective polypeptides within each subpanel. Chains within each subpanel correspond (from left to right) to chains B, D, F, H, J, and L (respectively) within each structure. Overlaid on the three subpanels in A is σA-weighted (2Fobs − Fcalc) difference electron density contoured at the 0.75 σ level and masked to within 2.5 Å of the side-chain atoms of PheB25 and its symmetry-related equivalents. The values displayed under the respective chains within the subpanels of B correspond to the side-chain occupancies of the PheB25 and its respective non-crystallographic symmetry equivalents within PDB code 1ZNJ. The side chain of NleB29 is not shown.
