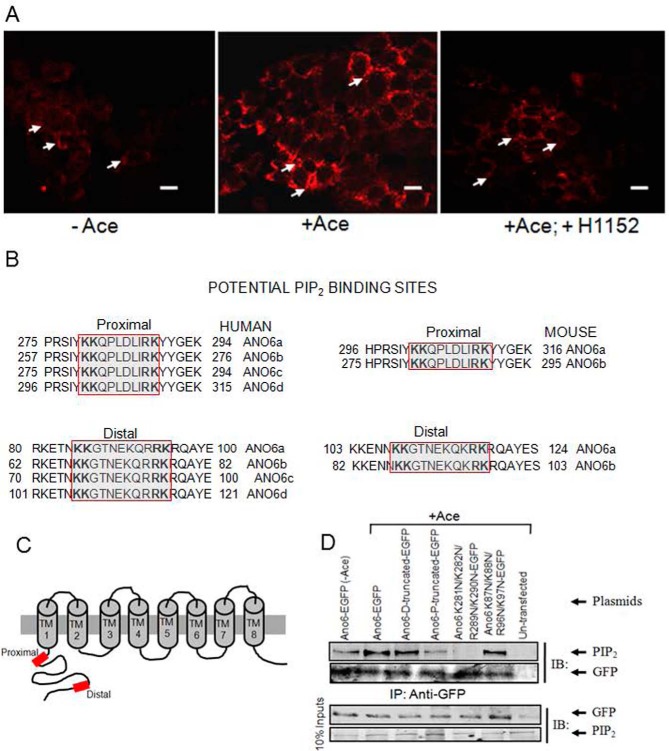
FIGURE 8.
Cellular abundance of PIP2 and its potential binding sites in the N terminus among the human and mouse ANO6 variants. A, Caco-2 wild-type cells were stimulated with Ace. Shown are representative images of fixed cells labeled for PIP2 in non-stimulated cells (control, left panel), Ace-stimulated cells (middle panel), and for cells that are pre-treated with H1152 and stimulated with Ace (right panel). Red color represents PIP2 abundance. Scale bars, 10 μm. B, sequence alignments at a comparable position in ANO6 variants showing two similar PIP2-binding motifs are conserved beginning immediately after the putative first membrane spanning domain of human (left) and mouse (right).Typical PIP2-binding motifs consisting of a cluster of basic residues are indicated by the red box. Sequences were obtained from the NCBI protein database. Accession numbers are as follows: variant 1 isoform a, NM_001025356.2 > NP_001020527.2; variant 2 isoform b, NM_001142678.1>NP_001136150.1; variant 3 isoform c, NM_001142679.1 > NP_001136151.1; and variant 4 isoform d, NM_001204803.1>NP_001191732.1. Mouse variant accession numbers are as follows: variant 1 isoform a, NM_001253813.1 > NP_0012407421.1, and variant 2 isoform b NM_175344.4 > NP_780553.2. C, schematic diagram illustrating the topology of human ANO6, highlighting two potential PIP2-binding sites (red box) of the channel N terminus. D, PIP2 co-immunoprecipitates with ANO6 in mANO6-GFP-transfected HEK293 cells whereas truncation mutants led to a significant decrease in the interaction. The cell lysates were incubated with anti-GFP-protein G-Sepharose beads, and inputs and immunoprecipitation eluate fractions were analyzed by Western blotting with either anti-PIP2 with or without Ace stimulation. PIP2 antibodies recognize GFP-tagged ANO6 protein in transfected HEK293 cells. No signal is detected in non-transfected HEK293 cells. This experiment was performed twice. The GFP blot for this experiment was presented to indicate the level of ANO6-GFP precipitation. D and P in truncated protein designate the deletion of distal and both distal and proximal KR motifs. Introduction of four mutations in the proximal and distal KR motifs are as indicated. IB, immunoblot; IP, immunoprecipitation.