Abstract
Mediator plays an integral role in activation of RNA polymerase II (Pol II) transcription. A key step in activation is binding of Mediator to Pol II to form the Mediator-Pol II holoenzyme. Here, we exploit a combination of biochemistry and macromolecular EM to investigate holoenzyme assembly. We identify a subset of human Mediator head module subunits that bind Pol II independent of other subunits and thus probably contribute to a major Pol II binding site. In addition, we show that binding of human Mediator to Pol II depends on the integrity of a conserved “hinge” in the middle module MED21-MED7 heterodimer. Point mutations in the hinge region leave core Mediator intact but lead to increased disorder of the middle module and markedly reduced affinity for Pol II. These findings highlight the importance of Mediator conformation for holoenzyme assembly.
Keywords: general transcription factor (GTF), RNA polymerase II, transcription, transcription coregulator, transcription regulation, Mediator
Introduction
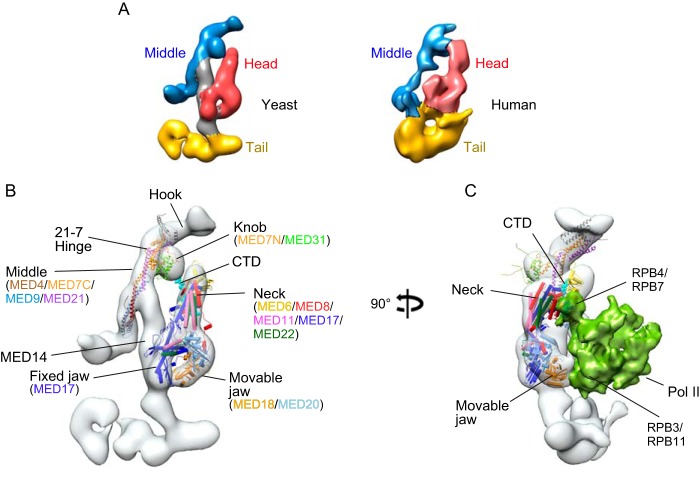
The multisubunit Mediator complex is an evolutionarily conserved RNA polymerase II (Pol II)3 coregulator. Mediator governs transcription at least in part by serving as a link between Pol II and transcription factors that regulate its activity during both initiation and elongation (1–5). Striking features of Mediator are its large size and architectural complexity. The simplest form of Mediator is the “core” complex, which is composed of more than 20 distinct polypeptides. Mediator core is organized into three modules (head, middle, and tail) arranged around MED14, which forms a central scaffold or stalk that supports intermodule interactions (Fig. 1, A and B) (6–8). In human cells, the Mediator head module includes seven highly conserved subunits (MED6, -8, -11, -17, -18, -20, and -22) as well a number of additional metazoan-specific subunits. The middle module includes eight well conserved subunits (MED1, -4, -7, -9, -19, -10, -21, and -31). Tail module subunits are the most evolutionarily divergent, and their number varies across species, perhaps reflecting their role as binding targets for activation domains of DNA binding transcription factors.
FIGURE 1.
A, comparison of yeast and human Mediator architecture. Left, ∼18 Å cryo-EM map of S. cerevisiae core Mediator; right, ∼30 Å map of human core Mediator, calculated from images of single particles preserved in stain (6). Positions of the middle, head, and tail modules are shown in blue, red, and yellow, respectively. B, yeast Mediator core complex; an X-ray structure of the yeast head module bound to CTD peptide (22) and a model of the middle module, based on partial X-ray structures and protein cross-linking (34), docked into the yeast cryo-EM map. C, structural model of the yeast Mediator-Pol II holoenzyme based on cryo-EM maps of a Mediator-PIC complex (17) and of a core yeast Mediator-ITC (18). Pol II subunits Rpb3/Rpb11 contact the movable jaw of Mediator, and the CTD (CTD peptide in cyan) is expected to bind between the head and the knob.
In addition to core Mediator, eukaryotes contain at least two other higher order Mediator-containing complexes (3–5, 9–11). One is the Mediator-Pol II holoenzyme, a stable complex of core Mediator and Pol II. The other is a complex composed of core Mediator and a cyclin-dependent kinase module, which in yeast is composed of CDK8, cyclin C, and two additional proteins designated MED12 and MED13. In metazoa, the Mediator-Pol II holoenzyme includes an additional subunit designated MED26, and the kinase module is assembled combinatorially from cyclin C, one of two cyclin-dependent kinases CDK8 or CDK19, and one of two MED12 and MED13 paralogs.
Substantial evidence argues that a key step in Mediator-dependent Pol II transcription is assembly of core Mediator with Pol II to form the holoenzyme (1, 8, 11, 12). Although the precise mechanisms and roles of specific Mediator subunits in formation of the holoenzyme are poorly understood, evidence from a variety of biochemical and EM structural studies argues that it involves binding of Pol II to specific surfaces on the Mediator head and middle modules (Fig. 1C) (6, 8, 13–18). In this report, we describe experiments investigating assembly of the human holoenzyme. Our findings define a subset of human Mediator head module subunits that contribute to formation of a Pol II binding surface. In addition, they bring to light a role for the hinge region of the middle module MED21-MED7 heterodimer in establishing a Mediator conformation permissive for Pol II binding.
Results
Identification of Mediator Head Module Subunits That Form an Interface with Pol II
We observed that treatment of cells with siRNA specific for MED21 led to disruption of the human Mediator-Pol II holoenzyme. Integrity of the holoenzyme was assessed following anti-FLAG immunopurification from nuclear extracts of a HeLa cell line stably expressing Pol II subunit RPB9 with an N-terminal FLAG epitope tag. In these experiments, siRNA treatment reproducibly reduced MED21 levels by 70–80% as estimated by densitometry of X-ray films exposed to chemiluminescent blots (Fig. 2A); this reduction in MED21 levels results in substantial inhibition of transcription of TNFα-induced activation of an NF-κB-driven reporter gene (Fig. 2B), indicating that activation of Pol II transcription is compromised under these conditions.
FIGURE 2.
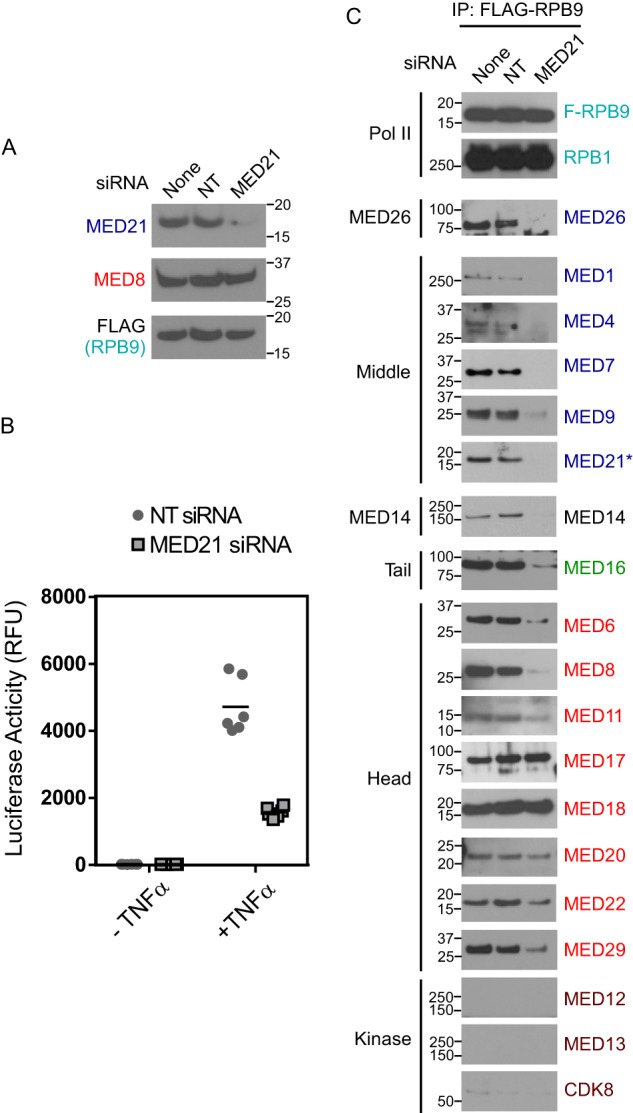
MED21 depletion disrupts the interaction of Mediator complex and RNA polymerase II. A, anti-MED21 immunoblot of lysates from untreated or siRNA-treated HeLa S3 cells from one of three biological replicates. 200 μg of lysate protein from untreated cells (none) or cells treated with non-targeting (NT) control or MED21 siRNA were applied to each lane. None, no siRNA; NT, non-targeting control siRNA; MED21, MED21 siRNA. In this and subsequent figures, positions of molecular mass markers (kDa) are indicated at the side of each immunoblot. B, effect of MED21 depletion on gene activation. Firefly luciferase activity was measured in lysates from HeLa cells with a stably integrated, NF-κB-responsive luciferase reporter, transfected with non-targeting control siRNA or MED21 siRNA. ● and ■, luciferase activity detected in lysates of cells treated with non-targeting or MED21 siRNA, respectively, from three independent experiments, each performed in duplicate. Horizontal lines, median. C, effect of MED21 depletion on Mediator-Pol II interactions. HeLa S3 cells stably expressing FLAG-RPB9 were transfected with non-targeting or MED21 siRNA, and Pol II-associated proteins were detected by immunoblotting of anti-FLAG immunoprecipitates (IP) using the indicated antibodies. Middle, Tail, and Head, Mediator modules to which individual subunits have been assigned.
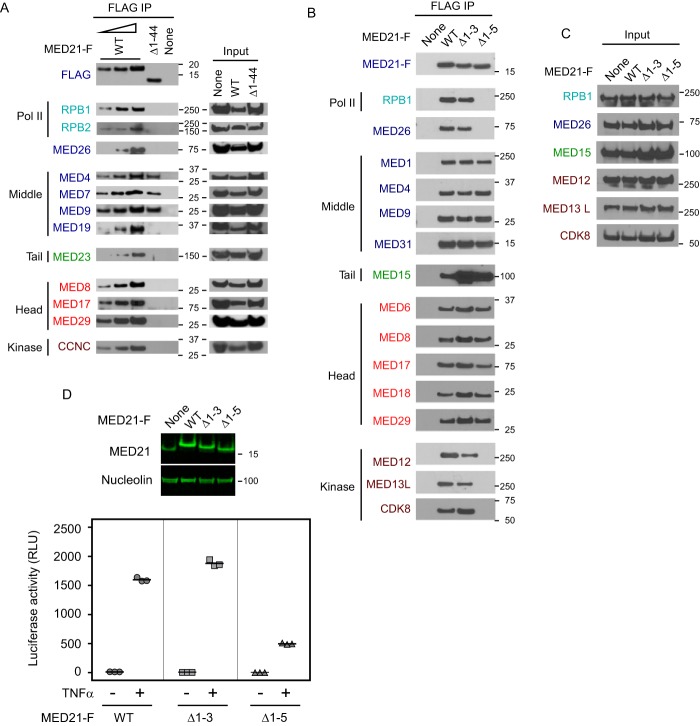
Representative subunits from the head, middle, and tail modules of human Mediator were detected by immunoblotting in FLAG immunoprecipitates from cells stably expressing FLAG-RPB9 (Fig. 2C). After MED21 depletion, there was a dramatic reduction in association of Pol II with many Mediator subunits. These included all middle and tail subunits tested as well as head module subunits MED6, MED8, and, to a lesser extent, metazoan-specific MED29. However, MED21 depletion had much less effect on binding of Pol II to a subset of head module subunits. In particular, there was no detectable change in the amount of MED17, MED18, and MED20 recovered in FLAG-RPB9 immunoprecipitations from control or MED21-depleted cells, whereas MED11 and MED22 were only modestly reduced. These results indicate (i) that loss of MED21 leads to dissociation of these subunits from the remainder of core Mediator and (ii) that some or all of these Mediator subunits interact directly with Pol II and contribute to a Pol II binding surface on human Mediator.
Although detailed structural information about human Mediator is not yet available, results of our previous EM analyses and biochemical mapping of subunit-subunit interactions, together with EM data presented below, argue that the overall architectures of human and yeast Mediators are conserved (Fig. 1A) (6). The findings reported here are consistent with models for yeast Mediator-Pol II interactions and provide biochemical evidence supporting the proposal that there is substantial conservation of head module organization and the Pol II-Mediator interaction throughout evolution (6, 18, 19). X-ray structures of the head module of yeast Mediator show that MED11, MED17, and MED22 constitute the structural core of the Head, and MED18 and MED20 form the moveable jaw (16, 20). Analyses of yeast Mediator-Pol II holoenzyme (17, 21), a yeast core Mediator-minimal preinitiation complex (18), and the complete Mediator-preinitiation complex (17) by cryo-EM and protein cross-linking show interactions between the moveable jaw (MED18-MED20) and RPB3-RPB11 and between MED17 and RPB4-RPB7 (Fig. 1C). Our findings suggest that interaction of Pol II with the moveable jaw and structural core of the head module are preserved after disruption of the middle module in MED21-depleted cells. However, head module subunits MED6 and MED8, which along with MED17 contribute to a binding surface for the C-terminal domain (CTD) of the largest Pol II subunit on the head module neck (22), showed greatly reduced association with intact Pol II in the absence of MED21. Thus, contacts between MED6, MED8, and the CTD are compromised after disruption of the middle module by MED21 depletion, and they are evidently dispensable for Pol II binding to head module subunits.
Why little MED6 or MED8 binds Pol II after MED21 depletion remains to be determined. Previous structural or protein cross-linking studies of yeast or human Mediator and Mediator modules provide little insight. Both yeast and human MED6 and MED8 can be assembled into recombinant head modules, with no apparent dependence on interaction with MED21 or other non-head subunits (7, 8, 16, 23). Neither MED6 nor MED8 have been reported to cross-link to MED21 or other middle module subunits; however, binding of yeast MED6 and MED21 has been detected in split-ubiquitin and GST pull-down assays (24). Yeast MED6 was found to cross-link to MED14 in the core Mediator-minimal preinitiation complex (18). Although not required for formation of recombinant head module assemblies, it is conceivable that contacts between MED6 and MED21 or MED14 could contribute to stable binding of MED6 and MED8 to Mediator in cells.
A Conserved MED21 N-terminal Region Needed for Binding of Mediator to Pol II and Kinase Module
MED21 is among the Mediator subunits most conserved throughout evolution (25) and is essential for cell viability in yeast and mice (26–28). The MED21 N terminus is particularly well conserved. Highlighting its importance for MED21 function, deletion of the first 15 amino acids from MED21 is lethal in Saccharomyces cerevisiae (29), whereas deletion of the first 7 amino acids interferes with both transcriptional activation and repression (24). Notably, a chimeric MED21 in which the N-terminal 77 amino acids of yeast MED21 are replaced with the corresponding region of human MED21 is functional in S. cerevisiae (27).
A potential role for MED21 N-terminal sequences in assembly of the Mediator-Pol II holoenzyme was initially brought to light by our observation that an N-terminal FLAG tag on MED21 interferes with isolation of the holoenzyme. In these experiments, nuclear extracts prepared from HeLa cell lines stably expressing MED21 with either N-terminal or C-terminal FLAG tags were subjected to anti-FLAG-agarose chromatography, and MED21-associated proteins in anti-FLAG-agarose eluates were identified by multidimensional protein identification technology (MudPIT) mass spectrometry.
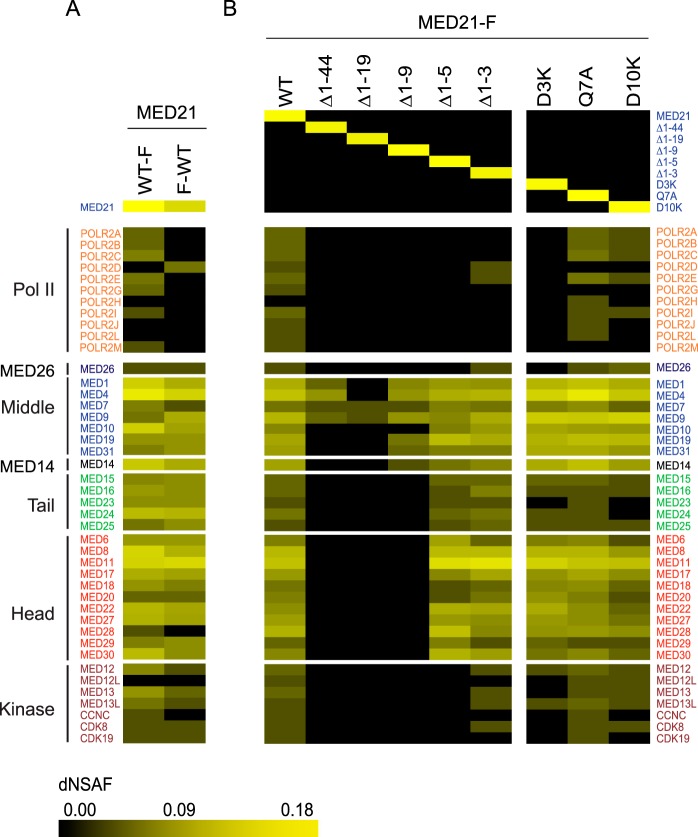
As shown in Fig. 3A, both N- and C-terminally tagged MED21 copurified with core Mediator, with subunits of the kinase module, and with the metazoan-specific subunit MED26. Whereas Pol II was readily detected in association with Mediator purified through C-terminally tagged MED21, little or no Pol II copurified with N-terminally FLAG-tagged MED21. These results indicate that the FLAG tag at the MED21 N terminus interferes with or is occluded by holoenzyme assembly.
FIGURE 3.
Identification of Mediator and Pol II subunits associated with full-length or mutant forms of MED21. Heat maps summarize estimates of relative recovery of Mediator and Pol II subunits after FLAG immunopurification through the indicated FLAG epitope-tagged MED21 protein from HeLa S3 (A) or FlpIN-TREx-293 (B) cell nuclear extracts, measured by MudPIT mass spectrometry and expressed as a normalized spectral abundance factor (dNSAF). In a MudPIT data set, the number of spectra derived from peptides of most proteins is a function of the protein's length and abundance. Thus, the relative amount of a particular protein in different samples can be estimated from dNSAF. Supporting MudPIT information is provided in supplemental Table S1. F-WT, N-terminally FLAG-tagged MED21; WT-F, C-terminally FLAG-tagged MED21. MED21-F, C-terminally FLAG-tagged WT MED21 or the indicated C-terminally tagged MED21 mutants. Mediator subunits were assigned to head, middle, tail, or kinase modules based on previously defined interactions and sequence similarity to yeast Mediator complex subunits (6, 25).
To explore the possibility that the conserved MED21 N terminus plays a role in assembly of the Mediator-Pol II holoenzyme, we generated additional human cell lines expressing a collection of N-terminal deletion mutants of MED21 with C-terminal FLAG tags. For these and subsequent experiments, wild type and mutant MED21 were expressed under control of a doxycycline-inducible promoter because we found in preliminary studies that constitutive expression of some of these mutants was toxic to cells.
As shown by MudPIT and by immunoblotting with antibodies directed against representative subunits of Pol II and each of the Mediator modules, MED21 deletion mutants lacking between 9 and 44 N-terminal amino acids were severely defective in assembly of both core Mediator and the Pol II holoenzyme (Figs. 3B and 4A). Although they were capable of interacting stably with a subset of middle module subunits, MED21 Δ1–44, MED21 Δ1–19, and MED21 Δ1–9 failed to assemble with significant amounts of head or tail module subunits, Pol II subunits, MED26, or the kinase module. In contrast, deletion of the first 5 MED21 amino acids had no apparent effect on the assembly of MED21 into core Mediator; however, it led to a dramatic loss of binding to Pol II (RPB1), MED26, and kinase module subunits (Figs. 3B and 4B), although expression of these proteins was the same in parental 293 cells and cells expressing wild type MED21 or MED21 N-terminal deletion mutants (Fig. 4C). Finally, a MED21 mutant lacking only the first 3 N-terminal amino acids bound core Mediator and readily detectable amounts of Pol II, MED26, and kinase module (Figs. 3B and 4B). To explore the effects of MED21 N-terminal deletions on gene expression in cells, we compared the extent of TNFα-induced activation of the NF-κB-driven reporter gene after doxycycline induction of wild type MED21, MED21 Δ1–3 (which assembles into Mediator that binds Pol II and kinase module), or MED21 Δ1–5 (which assembles into Mediator that fails to bind Pol II and kinase module). Suggesting that MED21 Δ1–5 acts as a dominant negative mutant, TNFα-induced reporter activity was substantially less in cells expressing MED21 Δ1–5 than in cells expressing wild type MED21 or MED21 Δ1–3.
FIGURE 4.
The MED21 N terminus contributes to Mediator assembly and is required for binding of Pol II and kinase module to Mediator core. Nuclear extracts from HeLa S3 (A) or Flp-In T-REx HEK293 (B) cells expressing either no exogenously expressed MED21 (None) or C-terminally FLAG-tagged WT or the indicated MED21 mutants were subjected to anti-FLAG M2 immunoaffinity purification and analyzed by immunoblotting using antibodies directed against Mediator or Pol II subunits. The first three lanes of A show 3-fold serial dilutions of immunopurified proteins from cells expressing wild type MED21-FLAG. C, immunoblots of cell lysates (input) used for FLAG immunoaffinity purification in B. D, MED21 lacking the first 5 amino acids is defective in TNFα-induced gene activation. Firefly luciferase activity was measured in lysates from Flp-InTM T-RExTM-293 cells stably expressing wild type or mutant MED21.
Point Mutations at the MED21-MED7 Hinge Region Alter the Interaction of Core Mediator with Pol II
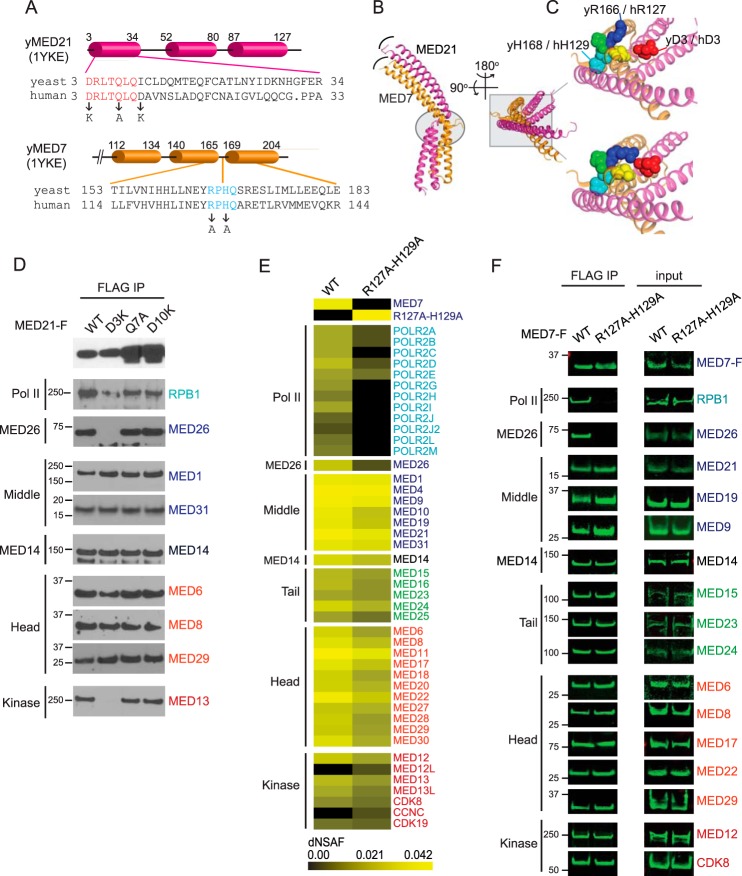
MED21 is a subunit of the Mediator middle module, where it forms a heterodimer with the highly conserved MED7 subunit (6, 7, 30). Like MED21, MED7 is essential for viability in yeast and metazoa (31, 32). Based on its X-ray structure, a heterodimer containing S. cerevisiae MED21 and the C-terminal domain of MED7 (MED7C) adopts a highly elongated, almost entirely α-helical structure (30). Although they have no sequence similarity, MED21 and MED7C are structurally similar, each being composed of three extended α-helices (Fig. 5, A–C). The MED21-MED7C heterodimer consists of two domains: a four-helix bundle formed by the two N-terminal α helices of each protein and a coiled-coil domain formed by their C-terminal helices. In the MED21-MED7C structure, the MED21 N terminus is located immediately adjacent to a conserved region of MED7 that contributes to a hinge-like structure located between the four-helix bundle and coiled-coil domains (30).
FIGURE 5.
MED21 and MED7 mutations in or near the MED21-MED7 hinge interfere with holoenzyme assembly. A, MED21 and MED7C secondary structure defined by X-ray crystallography of yeast MED21-MED7C heterodimer (Protein Data Bank entry 1YKE (30)) and sequence alignments of conserved MED21 N-terminal and MED7 hinge regions and positions of human MED21 and MED7 mutations. Cylinders, α-helices. B, superposition of two crystal forms of the yeast MED21 (magenta)-MED7C (orange) heterodimer, one containing chains B (MED21) and A (MED7C) and the other chains D (MED21) and C (MED7C), all from Protein Data Bank entry 1YKE. The four-helix bundles (which include α-helices 1 and 2 of MED21 and MED7C) from each crystal form are superimposed. Curved lines show difference in positions of the C-terminal α-helices of MED21 and MED7C in the two crystal forms. The MED21-MED7 hinge region is indicated by the oval. C, enlarged views of the region of the MED21-MED7C heterodimer outlined by the box in B, with the two crystal forms shown separately (top, chains D and C; bottom, chains B and A). Residues near the flexible hinge are shown as spheres. MED21 D3 (red) and MED7 residues Arg-166, Pro-167, His-168, and Gln-169 are blue, green, cyan, and yellow, respectively; in the more closed form of the MED21-MED7 heterodimer, MED21 Asp-3 is closer to Arg-166 of yeast MED7. y, yeast; h, human. D, nuclear extracts from Flp-In T-REx HEK293 cells expressing C-terminally FLAG-tagged WT or the indicated MED21 mutants were subjected to anti-FLAG M2 immunoaffinity purification and analyzed by immunoblotting using antibodies directed against Mediator or Pol II subunits. E, heat map showing number of spectra detected for Mediator and Pol II subunits after FLAG immunopurification through FLAG epitope-tagged wild type or mutant MED7 protein from HeLa S3 cell nuclear extracts, measured by MudPIT. Supporting MudPIT data are shown in supplemental Table S2. F, immunoblots showing Mediator and Pol II subunits that co-purified with FLAG-tagged wild type or mutant MED7. Immunoblots were visualized using an Odyssey infrared imaging system (LI-COR). IP, immunoprecipitation.
Comparison of two crystal forms of the yeast MED21-MED7C heterodimer indicates that differences in the positioning of the four-helix bundle relative to the coiled-coil domain are accompanied by changes in a network of contacts between residues in the MED21 N terminus and highly conserved MED7 residues that are located in the proposed hinge region (30). Such contacts might be expected to contribute to rigidity of the MED21-MED7 heterodimer. Because of the extended length of the MED21-MED7 heterodimer and its central position in the middle module, these contacts might also be important for middle module conformation (6, 16, 30, 33, 34).
Consistent with the possibility that residues located in or near the MED21-MED7 hinge influence assembly of the Mediator-Pol II holoenzyme, we identified specific MED21 and MED7 point mutations that block or greatly diminish formation of the holoenzyme without disrupting core Mediator. As shown by both MudPIT mass spectrometry (Fig. 3B) and immunoblotting (Fig. 5D), mutation of MED21 Asp-3 to Lys greatly reduced binding of core Mediator to Pol II and MED26, whereas mutation of MED21 Gln-7 to Ala or Asp-10 to Lys had little effect on binding of core Mediator to Pol II, MED26, or the kinase module. In addition, core Mediator containing a human MED7 mutant in which two of the highly conserved hinge residues, Arg-127 and His-129 (corresponding to Arg-166 and His-168 of yeast MED7; Fig. 5, A–C), were mutated to alanine also exhibited substantially reduced binding to Pol II and MED26 (Fig. 5, E and F). Interestingly, whereas the MED21 D3K mutation, like MED21 N-terminal deletion MED21 Δ1–5, led to a substantial reduction in binding of core Mediator to the kinase module, the MED7 hinge mutation had only a minimal effect on binding to the kinase module.
EM Analysis of Mutant Mediator Complexes
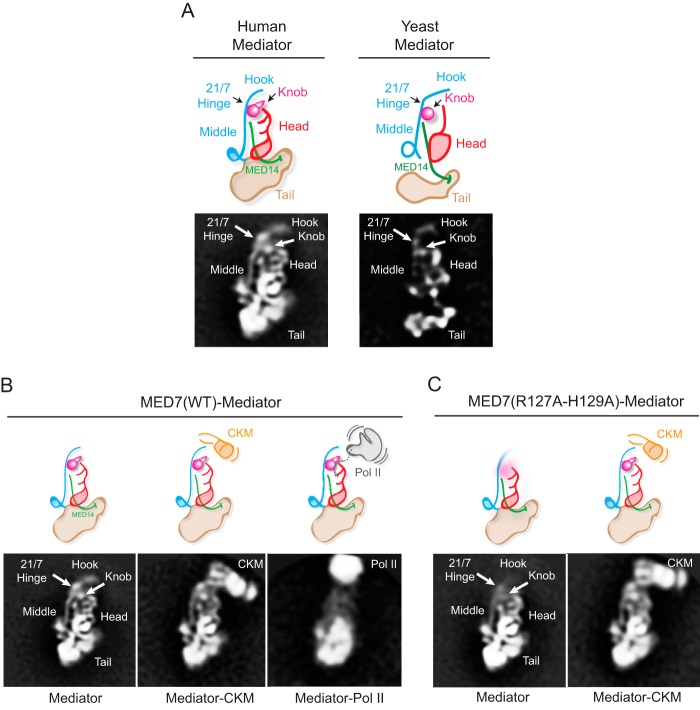
We used electron microscopy to explore the effects of mutating residues within the MED21-MED7 hinge region on the structure of human Mediator. To do so, Mediator was immunopurified from HeLa cell lines stably expressing FLAG-tagged versions of either wild type MED7 or the MED7 hinge mutant (R127A/H129A). Images of wild type and mutant Mediator complexes recorded after particle preservation in stain were used to calculate 2D image class averages showing the structure of the complexes in projection. Wild type core Mediator 2D class averages showed generally well ordered particles (Fig. 6, A and B, left) that appeared similar to averages of human core Mediator purified through the metazoan-specific subunit MED26 (6) or through a nuclear receptor bound to MED1 (35). Density resembling in size, shape, and position the head and middle modules, the MED19-containing “hook” region at the top of the Middle module, and the MED14 stalk can be readily identified by comparison with averages of Mediator from S. cerevisiae (6, 33) (Fig. 6A, right). Also, similar to previously published averages of human Mediator (35), density below the head and middle modules, in the position of the yeast tail module, can be seen; however, the human tail module appears larger and appears to have more connections with the lower portions of the middle and head modules than its yeast counterpart. Consistent with the results of the biochemical assays shown in Fig. 5, particles of a size and shape consistent with free core Mediator and core Mediator bound to the kinase module or to Pol II were observed (Fig. 6B).
FIGURE 6.
EM analysis of wild-type and MED7(R127A/H129A) human Mediators. A, a 2D class average obtained from stained particle images of wild-type human Mediator and a corresponding diagram summarizing the organization of the head, middle, and tail modules, the hook, and MED14 (left), based on comparison with wild-type yeast mediator (right). B, 2D class averages obtained after alignment and clustering of stained particle images from a wild-type human Mediator preparation. Free Mediator (left) and Mediator associated with kinase module (Mediator-CKM, center) or Pol II (Mediator-Pol II, right) were observed. The head, middle, and tail modules and the MED19-containing “hook” region at top of the middle module can be readily identified. The arrowhead marks the approximate position of the MED21-MED7 hinge region and the MED7(R127A-H129A) mutation, also inferred by comparison with the yeast Mediator structure (6). The Mediator-Pol II average is blurred by variability in the relative position of Mediator and Pol II, exacerbated by disruption of the complex after preservation in stain. C, class averages of free (left) and CKM-associated MED7(R127A/H129A) human Mediator. As illustrated in the diagrams, the MED7(R127A/H129A) mutation increases mobility of the hook and knob portions of the Middle module.
2D class averages calculated from images of human Mediator containing the MED7 hinge mutant (R127A/H129A) showed more heterogeneous particles. Nonetheless, clustering of images of the best ordered mutant MED7-Mediator particles resulted in averages that were generally similar to those obtained from wild type MED7-Mediator except that the hook and a structure resembling the S. cerevisiae Mediator knob, which includes the N-terminal portion of MED7 (MED7N) and MED31 (6), were not as well resolved as in wild-type human Mediator, presumably due to decreased stability and/or higher mobility. Interestingly, the hook and knob were well resolved in MED7(R127A/H129A)-Mediator that was bound to the kinase module, suggesting that these structures are stabilized upon binding of core Mediator to the kinase module. However, consistent with the biochemical results, no Pol II-bound Mediator particles were detected in preparations of mutant MED7-Mediator (Fig. 6C).
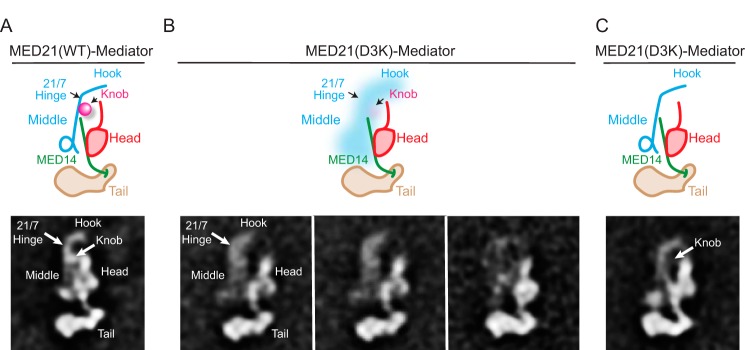
As noted above, expression of MED21(D3K) or other MED21 N-terminal mutants was quite toxic to human cells, and we were unable to prepare Mediator containing these mutants in sufficient quantity or quality for EM analysis. We were able, however, to purify S. cerevisiae MED21(D3K)-Mediator that was suitable for EM from a strain expressing C-terminally FLAG-tagged MED21(D3K). EM analysis confirmed that yeast Mediator containing wild type MED21-FLAG is indistinguishable from yeast Mediator purified through other subunits (Fig. 7A). MudPIT mass spectrometry of yeast MED21(D3K)-Mediator identified all core subunits but little Pol II or kinase module (supplemental Table S3). As was the case for the human Mediator containing the MED7 hinge mutant, EM analysis of yeast Mediator containing the MED21(D3K) mutant showed more heterogeneous particles than wild type Mediator. However, clustering of images of the best ordered particles resulted in 2D class averages in which the head, middle, and tail modules retained their general shapes and relative arrangement but in which the position of the middle module relative to the head and tail modules appeared more variable than in wild type Mediator (Fig. 7B). Whereas the apparent contact between the center of the middle module and the top of the central MED14 stalk was maintained in all yeast MED21(D3K)-Mediator class averages, contacts between the upper portion of the head module, corresponding to the head's neck, and the middle module appeared to be substantially disrupted. Increased variability in the position of the middle was also reflected by the appearance of individual averages in which the middle appeared less resolved than in wild type Mediator averages. In particular, density corresponding to the MED7N-MED31 knob was reduced or absent in most yeast MED21(D3K)-Mediator averages, although these subunits were still present in the mutant Mediator preparations (Fig. 7C and supplemental Table S3). This suggests that the interaction of the knob with the rest of the middle module is destabilized by the MED21(D3K) mutation.
FIGURE 7.
EM analysis of wild-type and MED21(D3K) yeast Mediators. A, a 2D class average from stained images and corresponding diagram showing that the structure and arrangement of head, middle, and tail modules is unaffected by introduction of a FLAG tag at the MED21 C terminus. The approximate position of the D3K mutation is indicated. B, 2D image class averages and corresponding diagrams showing variability in the position of the middle module in yeast Mediator containing MED21(D3K). C, a MED21(D3K) yeast Mediator class average in which the Middle module is well resolved shows that density corresponding to the MED7N-MED31 “knob” is absent. This agrees with what seems to be comparatively weak MED7N-MED31 density in the averages shown in B.
Discussion
In this report, we present findings that shed new light on the mechanism of assembly of the human Mediator-Pol II holoenzyme. In particular, we identify a collection of human Mediator subunits with roles in Pol II binding.
In one line of investigation, we identify a subset of human Mediator head module subunits that bind Pol II in the absence of other Mediator subunits and probably participate in the formation of a Pol II binding site on the surface of human Mediator; these include the moveable jaw subunits MED18 and MED20 as well as MED17, MED11, and MED22, which form the structural core of the head module. These observations raise the possibility that these subunits form a subcomplex within the human head module. As discussed earlier, these findings are consistent with results of prior protein cross-linking studies that identified contacts between yeast Pol II and subunits in the moveable jaw and structural core of the yeast head module (17, 18, 21), and they provide further evidence for conservation of Mediator structure and function from yeast to human.
Notably, we found that two additional head module subunits, MED6 and MED8, failed to bind Pol II under the same conditions. This finding was somewhat unexpected, because (i) the Pol II CTD is critical for binding of Pol II to Mediator (17) and (ii) an X-ray crystal structure of the yeast head module in complex with a peptide consisting of five copies of the CTD heptapeptide repeat demonstrated that CTD repeats contact four CTD-interacting regions (CIRs) in the head module consisting of portions of MED17, MED6, and MED8 (22). The first, CIR1, is composed entirely of helices from MED17. CIR2 includes surfaces from MED17 and MED8, CIR3 from MED8 and MED6, and CIR4 from MED6 alone. Our evidence that MED6 and MED8 are dispensable for binding of Pol II to the moveable jaw and structural core of the human head module suggests either (i) that CIR1-CTD contacts are most critical for the interaction of the Pol II CTD with human head module subunits or (ii) that binding to the CTD is less crucial in the context of this head module subcomplex than in the context of intact Mediator.
In a second line of investigation, we demonstrate that point mutations or small deletions in the vicinity of the evolutionarily conserved hinge region of the MED21-MED7 heterodimer have dramatic effects on the ability of Mediator to bind Pol II. Previous EM and cross-linking studies of yeast and human Mediator have established that the MED21-MED7 heterodimer is located in the middle module near the top of the MED14 stalk (6, 17, 18, 21). Based on existing models of yeast Mediator (6, 17, 18, 21), the MED21-MED7 hinge could, because of its location, affect the orientation or conformation of not only MED21-MED7, but also the MED7N-MED31-containing knob and the MED19-containing hook. As we have shown by EM, mutant Mediators containing either MED21 (D3K) or MED7 (R127A/H129A) exhibit increased disorder and/or flexibility of the knob and hook. We suggest that this increased disorder is due to increased flexibility of the MED21-MED7 heterodimer in the vicinity of the hinge. The acidic D3 residue at the MED21 N terminus is predicted to be in close proximity to Arg-127 of the MED7 hinge region (30); thus, it is reasonable to speculate that charge-charge repulsion between the MED21 (D3K) mutant and Arg-129 of MED7 could destabilize the hinge. Furthermore, the conserved histidine His-129 of the MED7 hinge region could be expected to establish a strong π-stacking interaction with either of two MED7 tyrosines in a way that could restrict movements of the MED7 helix; thus, it is possible that, by breaking those π-stacking interactions, mutation of MED7 His-129 to alanine could permit greater movement about the hinge region of MED21-MED7 and hence allow increased mobility of the upper portion of the middle module.
We found it surprising that mutant human Mediators containing either MED21(D3K) or MED7(R127A/H129A) failed to bind to Pol II, because they contained all Mediator subunits, including those head module subunits that we found can bind independently to Pol II when MED21 is depleted. These findings point to a model in which integrity of the MED21-MED7 hinge is required to establish a Mediator conformation permissive for binding Pol II. Mutations in the MED21-MED7 hinge region could affect Pol II binding by several, not mutually exclusive, mechanisms.
First, Tsai et al. (6) have obtained EM evidence that binding of Mediator to Pol II requires movement of Mediator modules with respect to one another as Mediator transitions from the core to holoenzyme form. Decreased rigidity of the middle module could make it such that the Mediator head and middle modules are most frequently oriented in a way that interferes with access of Pol II to the head's neck and jaw regions. Notably, MED21 residues in its second and third helices, which fall below and above the hinge, respectively, have been shown to cross-link to a small region within the MED17 N terminus in yeast Mediator (17, 18, 21). Increased movement about the MED21-MED7 hinge might weaken these contacts and thus lead to increased variability in the relative orientation of the middle and head modules.
Second, previous cross-linking studies have detected contacts between Pol II and the yeast middle module both above (MED19 and residues in MED21 helix 3) and below (MED4 and MED9) the MED21-MED7 hinge (17, 18, 21). Thus, it is possible that, in the context of the intact Mediator complex, a rigid middle module is needed to maintain the proper spatial orientation of these Pol II contact sites.
Finally, it has been proposed that interaction between core Mediator and the Pol II CTD might help trigger Mediator intermodule rearrangements necessary for holoenzyme formation (6). It is possible that increased disorder/mobility of the MED7N-MED31 knob caused by MED21-MED7 hinge mutations could affect Mediator CTD binding, because the knob is located directly across from the CTD binding site in the Mediator head module (Fig. 1, B and C) (6).
Experimental Procedures
Cell Culture
HeLa (ATCC CCL-2), HeLa S3 (ATCC CCL-2.2), and derivatives were maintained in adhesive culture in DMEM with 4.5 g/liter glucose, 10% (v/v) fetal bovine serum, 2 mm GlutaMAX, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 37 °C humidified incubator with 5% CO2. Culture medium for HeLa S3-mCAT1 derivatives expressing wild type or mutant MED21-FLAG, FLAG-RPB9, and wild type or mutant FLAG-MED7 was supplemented with 300 μg/ml hygromycin B and 100 μg/ml G418 sulfate. Culture medium for the HeLa-pGL4.32 reporter cell line (CCL2/4.32NF-κB-luc) was supplemented with 100 μg/ml hygromycin B. For large scale suspension cultures, HeLa S3 derivative cells were grown in spinner flasks with Joklik's modification of minimum essential medium Eagle (MEM) supplemented with 5% (v/v) calf serum, 2 mm GlutaMAX, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1× MEM non-essential amino acid solution (Thermo Fisher Scientific).
Cell lines derived from Flp-InTM T-RExTM 293 (Thermo Fisher Scientific) cells were cultured in the same medium as HeLa derivatives, except that it contained 10% (v/v) Tet System Approved fetal bovine serum (Clontech). Exogenously expressed MED21 wild type or mutants were induced in the Flp-InTM T-RExTM 293 stable cell lines by adding doxycycline at a final concentration of 2 μg/ml to the culture medium for 24 h.
All cells were confirmed to be free of mycoplasma contamination using the Lookout Mycoplasma Kit (MP0035, Sigma-Aldrich) or Universal Mycoplasma Detection Kit (30-1012K, ATCC).
Plasmid Construction and Generation of Stable Cell Lines
cDNAs encoding full-length human MED21 (AA449015) and MED7 (AI417176) were obtained as described (36). N-terminal deletion mutants of MED21 residues 1–44 (Δ1–44), 1–19 (Δ1–19), 1–5 (Δ1–5), and 1–3 (Δ1–3) were generated by PCR-based mutagenesis. MED7 double point mutation R127A/H129A and MED21 point mutation D3K, Q7A, or D10K were introduced by QuikChange II site-directed mutagenesis kit (Stratagene/Agilent). cDNAs encoding wild type and mutant human MED21 and MED7 with a FLAG tag at either their N or C termini were subcloned into pcDNA3.1/Hyg(−) (Thermo Fisher Scientific), into the retroviral vector pQCXIH (Clontech), or into pcDNA5/TO (Thermo Fisher Scientific) for tetracycline-regulated expression.
HeLa CCL2 cells were stably transfected with pGL4.32[luc2P/NF-κB-RE/Hygro] (Promega), which contains five copies of an NF-κB response element driving transcription of luc2P, to generate the NF-κB-responsive reporter cell line CCL2/4.32NF-κB-luc. pcDNA3.1/Hygro(−) derivatives encoding N- or C-terminally FLAG-tagged, wild type human MED21 were stably introduced into HeLa S3 cells using FuGene 6 (Promega), and hygromycin-resistant clones were isolated.
Clonal HeLa S3 cell lines stably transformed with mCAT-1 (murine cationic amino acids transporter), which functions as an ecotropic retrovirus receptor protein (mCAT-1) (37) and pQCXIH (Clontech) derivatives encoding N-terminally FLAG-tagged human RPB9 or C-terminally FLAG-tagged human MED21(Δ1–44), human MED7 wild type and double point mutant R127A-H129A were generated using the Plat-E system (38).
Tetracycline-inducible cell lines stably transfected with pcDNA5/TO encoding C-terminally FLAG-tagged MED21; MED21 point mutants D3K, Q7A, or D10K; and MED21 N-terminal deletion mutants Δ1–44, Δ1–19, Δ1–5, and Δ1–3 were generated in Flp-InTM T-RExTM-293 cells (Thermo Fisher Scientific) according to the manufacturer's instructions.
siRNA-mediated Gene Knockdown
All siRNA transfections were done using Lipofectamine RNAiMax (Thermo Fisher Scientific) according to the manufacturer's instructions. Exponentially growing HeLa S3 cells or CCL2/4.32NF-κB-luc reporter cells were seeded into 10-cm cell culture dishes or 6-well plates at a density of 3 × 106 cells/dish or 2.5 × 105 cells/well the day before the transfection. Cells were transfected with final 12 nm siRNA for human MED21 (J-016221-08), and control non-target pool (D-001810) obtained from GE Dharmacon.
Human MED21 (J-016221-08) and control non-target pool (D-001810) siRNAs (ON-TARGETplus, GE Dharmacon, final concentration 12 nm) were mixed with Opti-MEM I medium (Thermo Fisher Scientific) and transfected into HeLa S3 cells stably expressing FLAG-RPB9 or into the NF-κB-responsive CCL2/4.32NF-κB-luc reporter cell line using Lipofectamine RNAiMax (Thermo Fisher Scientific) according to the manufacturer's instructions. 24 h after transfection, the medium was replaced by fresh culture medium without siRNA. 48 h after siRNA transfection, cells were harvested, and whole cell extracts were prepared for anti-FLAG immunoprecipitation or reporter assay. We typically recovered 7–8 × 106 cells from each 10-cm dish. To prepare whole cell extracts, cells were resuspended in 400 μl of lysis buffer (50 mm HEPES-KOH (pH 7.9), 300 mm NaCl, 5 mm MgCl2, 0.2% Triton X-100, 20% glycerol, 1:200 (v/v) proteinase inhibitor mixture (P-8340, Sigma-Aldrich)) per 1.5 × 107 viable cells. Resuspended cells were vortexed vigorously for 1 min. After incubation for 15 min at 4 °C on a nutating shaker, whole cell lysates were collected by centrifugation at 20,817 relative centrifugal force for 20 min in a refrigerated microcentrifuge (Eppendorf). Protein concentration of the resulting extract was typically ∼15 mg/ml.
Reporter Assays
To assay the effect of siRNA-mediated depletion of MED21, 3 × 105 CCL2/4.32NF-κB-luc cells were plated in 6-well tissue culture plates the day before siRNA transfection and cultured in DMEM with high glucose supplemented with 10% charcoal-stripped fetal bovine serum, 2 mm GlutaMAX, and 1× MEM non-essential amino acids solution. 67 h after siRNA transfection, cells were treated with 20 nm human recombinant TNF-α (R&D Systems) or vehicle control at 37 °C for 5 h. To assay the effects of overexpressing wild type or mutant MED21 in cells, Flp-InTM T-RExTM-293 cells stably transformed with wild type or mutant MED21 were induced with tetracycline, plated in 6-well dishes as above, and transiently transfected with pGL4.32NF-κB-luc plasmid DNA (Promega). 24 h after transfection, cells were treated with 20 nm TNF-α or vehicle control for 5 h. Firefly luciferase activity was detected using the Dual-Luciferase reporter assay system (catalog no. E1910, Promega) with a VICTOR Light Luminescence counter (PerkinElmer Life Sciences).
Immunoblotting
Whole cell extracts or immunoaffinity-purified fractions were resolved in 4–12% gradient SDS-polyacrylamide NuPAGE gels (Thermo Fisher Scientific) or 8, 10, or 13% SDS-polyacrylamide gels and transferred onto a Hybond-P (GE Healthcare) or Immobilon-FL (EMD Millipore) polyvinylidene difluoride membrane using a Genie electrophoretic transfer system (Idea Scientific Co.). The following antibodies were used in Western blotting. Anti-FLAG M2 antibody (F3165), HRP-conjugated anti-mouse IgG (A3673), and HRP-conjugated anti-goat IgG (A4174) were from Sigma-Aldrich; HRP-conjugated anti-rabbit whole IgG (NA934) was from GE Healthcare; anti-Rpb2 (A300-349A), anti-MED1 (A300-793A), anti-CCNC (A301-989A), anti-MED12 (A300-774), anti-MED13 (A301–278A), anti-MED14 (A300-044A), anti-MED15 (A300-422A), anti-MED23 (A300-425A), and anti-MED24 (A300-472A) were from Bethyl Laboratories; anti-MED11 (ab79321), anti-MED13L (ab89831), anti-MED16 (ab28520), anti-MED26 (ab50619), anti-MED31 (ab54761), and anti-nucleolin (ab13541) were from AbCam; anti-RPB1 (N-20, sc-899), anti-CDK8 (C-19, sc-1521), anti-MED6 (sc-9433), and anti-MED8 (sc-81237) were from Santa Cruz Biotechnology; and anti-MED21 (H00009412-M05) and anti-MED20 (H00009477-B01P) were purchased from Novus Biologicals LLC. Anti-peptide antisera against a peptide corresponding to human CDK8 residues 397–414 or CDK19 residues 377–399 and antisera against full-length human MED4, MED7, MED9, MED17, MED18, MED22, and MED29 and mouse MED19 were raised in rabbits (Cocalico Biologicals, Inc.). IRDye800-conjugated anti-mouse IgG (610-131-121) and anti-rabbit IgG (611-131-122) were obtained from Rockland Immunochemicals Inc. Precision Plus ProteinTM all blue prestained protein standards were obtained from Bio-Rad. Protein signals were visualized either by chemiluminescent detection using SuperSignal West Dura extended duration substrate (catalog no. 34075, Thermo Fisher Scientific) or with an Odyssey fluorescence imaging system (LI-COR Biosciences).
Immunoaffinity Purification of Human Protein Complexes
Mediator or RNA polymerase II-associated protein complexes were purified from nuclear extracts prepared from cell lines stably expressing FLAG-tagged proteins using anti-FLAG M2-agarose (A2220, Sigma-Aldrich) as described previously (10, 39). Purified protein complexes were analyzed by immunoblotting and/or MudPIT (40).
Immunoaffinity purification from whole cell extracts of siRNA-treated cells was performed as described previously (6) with slight modifications. Briefly, based on total protein concentrations estimated using the Bradford protein assay (Bio-Rad), aliquots of whole cell extracts containing 5 mg of protein were precleared by incubating with 100 μl of packed Sepharose 4B for 15 min at 4 °C. To perform immunoaffinity purification, extracts were incubated for 4 h at 4 °C with 25 μl of packed anti-FLAG M2-agarose (A2220, Sigma-Aldrich). Beads were collected by centrifugation at 135 × g and then washed a total of five times by incubating for 5 min with 1 ml of FLAG washing buffer (50 mm HEPES-KOH, pH 7.9, 0.3 m NaCl, 10 mm KCl, 1.5 mm MgCl2 with 1:100 (v/v) protease inhibitor mixture (P8340, Sigma-Aldrich)) and collecting by centrifugation. Beads were eluted with total 50 μl of buffer containing 500 μg/ml FLAG peptide (F3290, Sigma-Aldrich), 50 mm HEPES-KOH, pH 7.9, 0.3 m NaCl, 10 mm KCl, 5% glycerol, with 1:100 (v/v) protease inhibitor mixture by incubating at 4 °C for 30 min or longer on the nutating mixer.
Yeast Mediator Purification
DNA encoding the yeast MED21 or MED21(D3K) ORFs with C-terminal FLAG and Protein A tags was introduced into the plasmid pRS305. Yeast strains expressing MED21-FLAG-Protein A or MED21(D3K)-FLAG-Protein A were established by recombination of the resulting pRS305 derivatives into the LEU2 locus of S. cerevisiae strain W303 (Table 1). Yeast were incubated overnight at 30 °C in 100 ml of synthetic drop-out medium, transferred to 8 liters of 2× YPD medium, and then grown to OD ∼3.0. Yeast cells were harvested, washed twice with double-deionized water, and frozen in liquid nitrogen. Frozen cells were broken by blending with dry ice and liquid nitrogen. Approximately 100 g of broken yeast cell powder were resuspended in 100 ml of buffer A (100 mm Tris-HCl, pH 8.0, 500 mm KOAc, 0.5 mm EDTA, 20% glycerol, 5 mm β-mercaptoethanol, and protease inhibitors) containing protease inhibitors (Sigma-Aldrich) at a final concentration of 20 μg/ml benzamidine hydrochloric hydrate (B6506), 1 μg/ml pepstatin A (P4265), 1 μg/ml leupeptin (L2884), and 1 mm phenylmethanesulfonyl fluoride (P7626). The resulting lysate was cleared by centrifugation. The supernatant was precipitated with 55% ammonium sulfate, and the resulting pellet was resuspended in buffer B (20 mm HEPES, pH 7.6, 300 mm KOAc, 0.5 mm EDTA, 10% glycerol, 0.05% Nonidet P-40, 5 mm β-mercaptoethanol), including fresh protease inhibitors. After the suspension was clarified by centrifugation, the supernatant was incubated at 4 °C for 2 h with 0.4 ml of IgG-Sepharose resin beads (GE Healthcare, catalog no. 17-0969). After incubation, the beads were washed extensively with buffer B, followed by further washing with buffer B plus 1 mm DTT. After washing, 100 units of AcTEVTM protease (Thermo Fisher Scientific, catalog no. 12575) were added to the resin beads and incubated overnight at 4 °C. Mediator complexes were eluted with three column volumes of buffer B plus 1 mm DTT.
TABLE 1.
List of yeast strains used in this study
| S. cerevisiae strain | Description | Reference/Source |
|---|---|---|
| W303 | MATa/MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15} [phi+] | Ref. 50 |
| Med21–3F-ProtA | leu2-3,112::LEU2-MED21(WT)-3XFLAG-ProteinA | This work |
| Med21(D3K)-3F-ProtA | leu2-3,112::LEU2-MED21 (D3K)-3XFLAG-ProteinA | This work |
MudPIT
Proteins were precipitated with TCA and solubilized in Tris-HCl, pH 8.5, and 8 m urea. Tris(2-carboxylethyl)-phosphine hydrochloride (Pierce) and chloroacetamide (Sigma) were added to a final concentration of 5 and 10 mm, respectively. Samples were digested overnight at 37 °C using endoproteinase Lys-C at 1:50 (w/w) (Roche Applied Science), brought to a final concentration of 2 m urea and 2 mm CaCl2, and incubated overnight at 37 °C with trypsin (Promega) at 1:100 (w/w). Formic acid (5% final) was added to stop the reactions. Peptide mixtures were loaded onto 100-μm fused silica microcapillary columns packed with 8 cm of Aqua C18 reverse phase resin (Phenomenex), 3 cm of Partisphere SCX 5 resin (Whatman), and 2 cm of Aqua C18 reverse phase resin (41). Loaded microcapillary columns were placed in-line with linear ion trap mass spectrometers (LTQ, Thermo Scientific) coupled to quaternary Agilent 1100 or 1200 series HPLCs, and fully automated 10-step chromatography runs (40) with dynamic exclusion enabled for 120 s were performed. MS/MS data sets were searched using SEQUEST (42). Samples from human cells were searched against a database of 111,379 sequences, consisting of 55,508 Homo sapiens non-redundant proteins (downloaded from NCBI on February 4, 2014): 9 MED21 mutant sequences; 177 common contaminants (including human keratins, IgGs, and proteolytic enzymes); and, to estimate false discovery rates, 55,685 randomized amino acid sequences derived from each non-redundant protein entry. Samples from S. cerevisiae were searched against a database of 12,038 sequences, consisting of 5843 S. cerevisiae non-redundant proteins (downloaded from NCBI on February 26, 2013), 177 contaminants, and 6019 randomized amino acid sequences derived from non-redundant protein entries. Peptide/spectrum matches were sorted, selected, and compared using DTASelect/CONTRAST (43). Proteins had to be detected by at least two spectra; average false discovery rates at the protein and spectral levels were 0.54 and 0.17%, respectively. Normalized spectral abundance factors (dNSAFs) were calculated as described (44).
Electron Microscopy
EM samples were prepared by applying purified Mediator particles (yeast wild type MED21-Mediator, yeast MED21(D3K)-Mediator, human wild type MED7-Mediator, or human MED7(R127A/H129A)-Mediator) to continuous carbon grids and preserving them by staining with 0.75% (w/v) uranyl formate. Particle images (at 52,000× magnification, 2.05 Å/pixel) were automatically acquired on a Tecnai Spirit electron microscope (FEI) equipped with a LaB6 filament and operating at an acceleration voltage of 120 kV, using the Leginon package (45). Images were recorded on a 4096 × 4096 TemCam F416 CMOS detector (Tietz, Inc.). Particle images for yeast Mediator were picked manually and aligned and classified after 2-fold pixel binning (final pixel size 4.1 Å/pixel) using the Iterative Stable Alignment and Clustering (ISAC) program implemented in the SPARX software package (46). Particle images for human mediator were picked using the Appion pipeline (47, 48), and the reference-free 2D class averages were calculated using RELION version 1.4 (49). A total of 7805 human MED7(WT)-Mediator images, 8582 human MED7(R127A/H129A)-Mediator images, 5662 wild type yeast Mediator images, and 3924 yeast MED21(D3K)-Mediator images were included in the respective EM data sets.
Original Data
Original data underlying this paper can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1103. Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
Author Contributions
S. S., C. T.-S., R. C. C., and J. W. C. conceived and designed the study and analyzed data; S. S. and C. T.-S. generated cell lines and yeast strains and were responsible for biochemical experiments; K.-L. T., X. D., and F. A. were responsible for generating and analyzing EM data; M. S., A. S., M. P. W., and L. F. were responsible for proteomic analyses; R. C. C., J. W. C., S. S., C. T.-S., and F. A. wrote the paper; and all authors contributed to its editing.
Supplementary Material
Acknowledgments
We thank Sue Jaspersen and Jennifer Gardner for advice and help with yeast manipulation; Andrew Paoletti for help generating the FLAG-Rpb9 expressing HeLa cell line; members of the Stowers Institute's Tissue Culture and Molecular Biology Core Facilities for maintaining cell lines and DNA sequencing, respectively; Quentin Michard for help with initial phases of this study; and Sara Jackson for help preparing the manuscript.
Note Added in Proof
There were several errors in the version of this article that was published as a Paper in Press on November 7, 2016. In Fig. 2C, the marker labels for the MED29 immunoblot were incorrect. In Fig. 4A, the wrong input blot for MED4 was used. These errors have now been corrected. The corrections do not alter the interpretation of the data.
This work was supported in part by a grant to the Stowers Institute from the Helen Nelson Medical Research Fund at the Greater Kansas City Community Foundation and by NIGMS, National Institutes of Health, Grants GM41628 (to R. C. C. and J. W. C.) and GM067167 (to F. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S3.
- Pol II
- RNA polymerase II
- CTD
- C-terminal domain
- CIR
- CTD-interacting region
- MEM
- minimum essential medium Eagle
- MudPIT
- multidimensional protein identification technology.
References
- 1. Kim Y. J., Björklund S., Li Y., Sayre M. H., and Kornberg R. D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 [DOI] [PubMed] [Google Scholar]
- 2. Myers L. C., and Kornberg R. D. (2000) Mediator of Transcriptional Regulation. Annu. Rev. Biochem. 69, 729–749 [DOI] [PubMed] [Google Scholar]
- 3. Malik S., and Roeder R. G. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conaway R. C., and Conaway J. W. (2013) The Mediator complex and transcription elongation. Biochim. Biophys. Acta 1829, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen B. L., and Taatjes D. J. (2015) The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai K. L., Tomomori-Sato C., Sato S., Conaway R. C., Conaway J. W., and Asturias F. J. (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang J. S., Kim S. H., Hwang M. S., Han S. J., Lee Y. C., and Kim Y. J. (2001) The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276, 42003–42010 [DOI] [PubMed] [Google Scholar]
- 8. Cevher M. A., Shi Y., Li D., Chait B. T., Malik S., and Roeder R. G. (2014) Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mittler G., Kremmer E., Timmers H. T., and Meisterernst M. (2001) Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., and Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 11. Näär A. M., Taatjes D. J., Zhai W., Nogales E., and Tjian R. (2002) Human CRSP interacts with RNA polymerase II and adopts a specific conformation. Genes Dev. 16, 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soutourina J., Wydau S., Ambroise Y., Boschiero C., and Werner M. (2011) Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331, 1451–1454 [DOI] [PubMed] [Google Scholar]
- 13. Dotson M. R., Yuan C. X., Roeder R. G., Myers L. C., Gustafsson C. M., Jiang Y. W., Li Y., Kornberg R. D., and Asturias F. J. (2000) Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 14307–14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis J. A., Takagi Y., Kornberg R. D., and Asturias F. A. (2002) Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 [DOI] [PubMed] [Google Scholar]
- 15. Cai G., Chaban Y. L., Imasaki T., Kovacs J. A., Calero G., Penczek P. A., Takagi Y., and Asturias F. J. (2012) Interaction of the Mediator head module with RNA polymerase II. Structure 20, 899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larivière L., Plaschka C., Seizl M., Wenzeck L., Kurth F., and Cramer P. (2012) Structure of the Mediator head module. Nature 492, 448–451 [DOI] [PubMed] [Google Scholar]
- 17. Robinson P. J., Trnka M. J., Bushnell D. A., Davis R. E., Mattei P. J., Burlingame A. L., and Kornberg R. D. (2016) Structure of a complete Mediator-RNA polymerase II pre-initiation complex. Cell 166, 1411–1422.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E. V., Borchers C. H., Baumeister W., Herzog F., Villa E., and Cramer P. (2015) Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 [DOI] [PubMed] [Google Scholar]
- 19. Asturias F. J., Jiang Y. W., Myers L. C., Gustafsson C. M., and Kornberg R. D. (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283, 985–987 [DOI] [PubMed] [Google Scholar]
- 20. Imasaki T., Calero G., Cai G., Tsai K. L., Yamada K., Cardelli F., Erdjument-Bromage H., Tempst P., Berger I., Kornberg G. L., Asturias F. J., Kornberg R. D., and Takagi Y. (2011) Architecture of the Mediator head module. Nature 475, 240–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson P. J., Trnka M. J., Pellarin R., Greenberg C. H., Bushnell D. A., Davis R., Burlingame A. L., Sali A., and Kornberg R. D. (2015) Molecular architecture of the yeast Mediator complex. Elife 10.7554/eLife.08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson P. J., Bushnell D. A., Trnka M. J., Burlingame A. L., and Kornberg R. D. (2012) Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 109, 17931–17935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takagi Y., Calero G., Komori H., Brown J. A., Ehrensberger A. H., Hudmon A., Asturias F., and Kornberg R. D. (2006) Head module control of mediator interactions. Mol. Cell 23, 355–364 [DOI] [PubMed] [Google Scholar]
- 24. Gromöller A., and Lehming N. (2000) Srb7p is a physical and physiological target of Tup1p. EMBO J. 19, 6845–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bourbon H. M. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hengartner C. J., Thompson C. M., Zhang J., Chao D. M., Liao S. M., Koleske A. J., Okamura S., and Young R. A. (1995) Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9, 897–910 [DOI] [PubMed] [Google Scholar]
- 27. Chao D. M., Gadbois E. L., Murray P. J., Anderson S. F., Sonu M. S., Parvin J. D., and Young R. A. (1996) A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature 380, 82–85 [DOI] [PubMed] [Google Scholar]
- 28. Tudor M., Murray P. J., Onufryk C., Jaenisch R., and Young R. A. (1999) Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13, 2365–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gromöller A., and Lehming N. (2000) Srb7p is essential for the activation of a subset of genes. FEBS Lett. 484, 48–54 [DOI] [PubMed] [Google Scholar]
- 30. Baumli S., Hoeppner S., and Cramer P. (2005) A conserved mediator hinge revealed in the structure of the MED7·MED21 (Med7·Srb7) heterodimer. J. Biol. Chem. 280, 18171–18178 [DOI] [PubMed] [Google Scholar]
- 31. Myers L. C., Gustafsson C. M., Bushnell D. A., Lui M., Erdjument-Bromage H., Tempst P., and Kornberg R. D. (1998) The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwon J. Y., Kim-Ha J., Lee B. J., and Lee J. (2001) The MED-7 transcriptional mediator encoded by let-49 is required for gonad and germ cell development in Caenorhabditis elegans. FEBS Lett. 508, 305–308 [DOI] [PubMed] [Google Scholar]
- 33. Wang X., Sun Q., Ding Z., Ji J., Wang J., Kong X., Yang J., and Cai G. (2014) Redefining the modular organization of the core Mediator complex. Cell Res. 24, 796–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larivière L., Plaschka C., Seizl M., Petrotchenko E. V., Wenzeck L., Borchers C. H., and Cramer P. (2013) Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res. 41, 9266–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsai K. L., Sato S., Tomomori-Sato C., Conaway R. C., Conaway J. W., and Asturias F. J. (2013) A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato S., Tomomori-Sato C., Banks C. A., Sorokina I., Parmely T. J., Kong S. E., Jin J., Cai Y., Lane W. S., Brower C. S., Conaway R. C., and Conaway J. W. (2003) Identification of mammalian Mediator subunits with similarities to yeast Mediator subunits Srb5, Srb6, Med11, and Rox3. J. Biol. Chem. 278, 15123–15127 [DOI] [PubMed] [Google Scholar]
- 37. Albritton L. M., Tseng L., Scadden D., and Cunningham J. M. (1989) A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57, 659–666 [DOI] [PubMed] [Google Scholar]
- 38. Morita S., Kojima T., and Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 39. Tomomori-Sato C., Sato S., Conaway R. C., and Conaway J. W. (2013) Immunoaffinity purification of protein complexes from Mammalian cells. Methods Mol. Biol. 977, 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Florens L., and Washburn M. P. (2006) Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328, 159–175 [DOI] [PubMed] [Google Scholar]
- 41. McDonald W. H., Ohi R., Miyamoto D. T., Mitchison T. J., and Yates J. R. (2002) Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219, 245–251 [Google Scholar]
- 42. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 43. Tabb D. L., McDonald W. H., Yates J. R. 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y., Wen Z., Washburn M. P., and Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 45. Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C. S., and Carragher B. (2005) Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 [DOI] [PubMed] [Google Scholar]
- 46. Hohn M., Tang G., Goodyear G., Baldwin P. R., Huang Z., Penczek P. A., Yang C., Glaeser R. M., Adams P. D., and Ludtke S. J. (2007) SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 [DOI] [PubMed] [Google Scholar]
- 47. Lander G. C., Stagg S. M., Voss N. R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P. W., Lyumkis D., Potter C. S., and Carragher B. (2009) Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Voss N. R., Yoshioka C. K., Radermacher M., Potter C. S., and Carragher B. (2009) DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scheres S. H. (2012) A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rothstein R. J. (1983) One-step gene disruption in yeast. Methods Enzymol. 101, 202–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.