Figure 8. The NED and NIM are functionally equivalent.
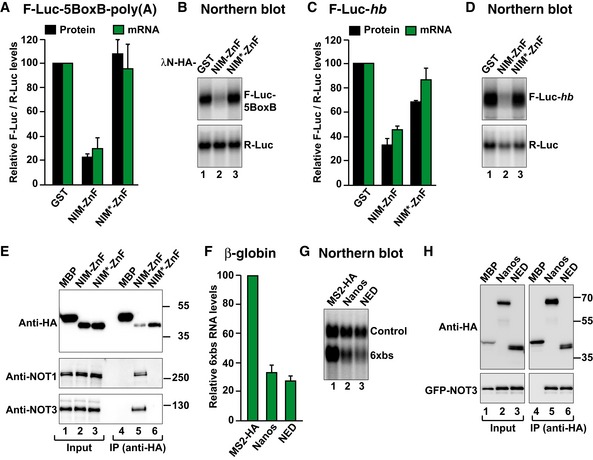
- Tethering assay using a chimeric Nanos protein and the F‐Luc‐5BoxB reporter in S2 cells. The chimeric Nanos protein contains the NIM of human Nanos2 (either wild type (NIM‐ZnF) or mutated (NIM*‐ZnF)), fused to the Dm ZnF domain. A plasmid expressing GST served as a negative control. F‐Luc activity and mRNA levels were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (A).
- The activity of GST‐HA‐tagged Nanos chimeric protein was tested in S2 cells expressing an F‐Luc‐hb reporter. A plasmid expressing GST served as a negative control. F‐Luc activity and mRNA levels were analyzed as described in Fig 1B. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (C).
- Western blot analysis showing the interaction of the HA‐tagged Nanos chimeric protein (NIM‐ZnF or NIM*‐ZnF) with endogenous Dm NOT1 and NOT3. HA‐MBP served as a negative control.
- Tethering assays in human HEK293T cells, using a β‐globin reporter containing 6 binding sites (6xbs) for the MS2 protein and MS2‐HA‐tagged Dm Nanos or NED fragment. A plasmid expressing an mRNA lacking MS2 binding sites (Control) served as a transfection control. The β‐globin‐6xbs mRNA levels were normalized to those of the control mRNA and set to 100 in the presence of MS2‐HA. The panel shows mean values ± standard deviations from three independent experiments.
- Northern blot of representative RNA samples corresponding to the experiment shown in (F).
- Co‐immunoprecipitation assay in human HEK293T cells showing the interaction of the HA‐tagged Dm Nanos and the NED fragment with GFP‐tagged NOT3 in human cells. HA‐MBP served as a negative control. The inputs (1%) and bound fractions (10% of HA‐tagged proteins and 30% of GFP‐NOT3) were analyzed by Western blotting.
Source data are available online for this figure.
