Figure 4.
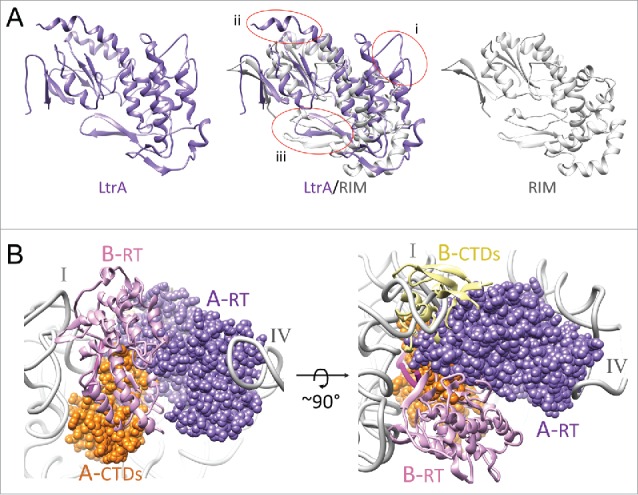
Differences between LtrA and RIM. (A) Comparison of LtrA and RIM structures. The RT domains of LtrA (PDB ID: 5g2x, purple) and RIM (PDB ID: 5hhj, gray) are shown as individual and superimposed ribbons. Three disparate regions between the 2 structures are circled (i – iii). LtrA is larger than RIM, with insertions in (i) the NTE and RT3 motif (encompassing the IFD), (ii) the 4a region (a significant portion of the 4a region that extended beyond the α-helix 5 was disordered in the RNP structure and could not be modeled), and (iii) the RT1/RT2 motif with a β-sheet hairpin, which is differently oriented between the structures. (B) Probing LtrA dimer formation. LtrA is shown in dimeric configuration of the RT domains RT0-RT7 as observed in the X-ray crystallographic study of RIM. The RNA-bound copy of the LtrA (chain A), from the cryo-EM study, is shown as a space-filled model, whereas the second superimposed copy of LtrA (chain B) is shown as ribbons. Labels: A-RT (dark purple) and B-RT (light purple) depict NTE and maturase domains of chains A and B, respectively; A-CTDs (orange) and B-CTDs (yellow) depict the thumb, DNA-binding, and endonuclease domains of chains A and B, respectively. I and IV, indicate the 2 LtrA-interacting domains of the RNA intron (gray).
