ABSTRACT
EF-G, EF4, and BipA are members of the translation factor family of GTPases with a common ribosome binding mode and GTPase activation mechanism. However, topological variations of shared as well as unique domains ensure different roles played by these proteins during translation. Recent X-ray crystallography and cryo-electron microscopy studies have revealed the structural basis for the involvement of EF-G domain IV in securing the movement of tRNAs and mRNA during translocation as well as revealing how the unique C-terminal domains of EF4 and BipA interact with the ribosome and tRNAs contributing to the regulation of translation under certain conditions. EF-G, EF-4, and BipA are intriguing examples of structural variations on a common theme that results in diverse behavior and function. Structural studies of translational GTPase factors have been greatly facilitated by the use of antibiotics, which have revealed their mechanism of action.
KEYWORDS: BipA, EF-G, EF4, ribosome, translocation, trGTPase
Introduction
In all cells, proteins are synthesized based on mRNA templates via amino acid charged tRNAs by ribosomes; macromolecular RNA-protein assemblies composed of 2 unequally sized subunits. The bacterial ribosome (70S) larger subunit (50S) consists of 2 RNA molecules (23S and 5S rRNAs) and 33 L-proteins (prefix “L” for large) whereas the smaller subunit (30S) consists of one RNA molecule (16S rRNA) and 21 S-proteins (prefix “S” for small). All ribosomal components are present in one copy, except for the L12 stalk proteins (L12 and its N-acetylated form L7) present in 4 to 6 copies per bacterial ribosome.1 The ribosome harbors 3 tRNA binding sites: the A (aminoacyl-tRNA), P (peptidyl-tRNA), and E (exiting tRNA) sites. It functions by oscillating between 2 main states as it moves along an mRNA decoding consecutive codons, namely the pre-translocational (PRE) and post-translocational (POST) states with tRNAs either in A and P or P and E sites, respectively.
Translational guanosine triphosphatase factors (trGTPases) are proteins with ribosome-dependent GTPase activity that occupy a characteristic position in the ribosome and are defined by the presence of highly conserved GTPase (G) domain. Protein synthesis in bacteria involves 4 main trGTPase factors: initiation factor 2 (IF2), elongation factors Tu and G (EF-Tu and EF-G), and release factor 3 (RF3). Thus, trGTPase factors function in all the major phases of translation: initiation, elongation (both mRNA decoding and mRNA-tRNAs complex translocation), termination, and recycling in a guanosine triphosphate (GTP)-dependent manner. Several additional GTPase factors, including elongation factor 4 (EF4, previously known as LepA), and BipA (also known as TypA and YihK) have been revealed to be associated with ribosomes under certain conditions.
EF4 and BipA are paralogs of EF-G, yet, regardless of their structural similarity, exhibit distinct functions that can be attributed to their varied domain arrangements and accessory domains with unique ribosome and tRNA binding modes. In this review, we discuss the structural and functional similarity as well as diversity of these 3 ribosome-dependent GTPase factors in light of recent X-ray crystallography and cryo-electron microscopy (cryo-EM) studies starting with a brief introduction to EF-G, EF4, and BipA.
EF-G
EF-G is responsible for ensuring the rapid and coordinated movement of mRNA along with bound tRNAs through the ribosome at the end of each round of polypeptide elongation, a process known as translocation. In previous models it was believed that movement of tRNAs on the 50S subunit (hosting the peptidyl-transferase center, PTC) from A and P to P and E sites, creating A/P and P/E hybrid site tRNAs, respectively, accompanied by subunit rotation relative to one another (also known as the ratcheting movement) occurs spontaneously,2-4 while the movement of codon-anticodon duplexes on the 30S subunit (hosting the decoding center, DC) from A and P (PRE state) to P and E sites (POST state), respectively, accompanied by 30S head swiveling, was believed to require GTP hydrolysis by EF-G.2 However, kinetic studies5,6 now suggest that tRNAs move synchronously on the 2 ribosomal subunits mediated by EF-G and GTP hydrolysis and that EF-G binding greatly accelerates the formation of A/P and P/E hybrid state. In contrast, according to a time-resolved EM study, the 2 tRNAs move in a non-synchronous step-by-step manner as revealed by the presence of an intermediate state with tRNAs in A/A and hybrid P/E sites.7 Rodnina and co-workers conclude in a recent review that EF-G facilitates tRNA movement by a combination of functioning as the pawl in a Brownian ratcheting device and as a power stroke mechanism.8 EF-G-driven hydrolysis of GTP ultimately leads to stabilization of the mRNA-tRNAs complex 3 nucleotides downstream of the ribosome, as well as restoring the ribosome to the classical unrotated state.3,9-11 The guanosine diphosphate (GDP)-bound EF-G molecule then dissociates from the ribosome leaving the A site empty (POST complex) and ready to accept the next aminoacyl-tRNA delivered by EF-Tu. Once the synthesis of the peptide chain is completed upon translation termination factors encountering a stop codon and triggering the release of the nascent peptide chain, the ribosome is left with mRNA and a deacylated tRNA in either P or P/E hybrid site. This complex is dissociated into free subunits, which can be used in subsequent rounds of translation initiation by ribosome recycling factor (RRF) together with EF-G,12 a process known as ribosome recycling. The dissociated subunits can be recycled in subsequent rounds of translation initiation.
EF-G is the only classical trGTPase that functions in 2 different phases of protein synthesis. Unfortunately, there is no detailed understanding on how EF-G facilitates ribosome recycling, as the atomic structure of the ribosome in complex with both RRF and EF-G has not been characterized yet. In contrast, both X-ray crystallography and cryo-EM studies have generated ample structural data on the EF-G/ribosome interaction during various stages of translocation, offering insight into how GTP hydrolysis is triggered by the ribosome and is coordinated with the conformational rearrangements required for translocation (discussed in more detail in following sections).
EF4
Unlike the universally conserved EF-G (eEF2 in eukaryotes), its paralog EF4 is ubiquitously conserved in bacterial (only known exceptions are Streptococcus pyogenes and Carsonella ruddii) as well as mitochondrial and chloroplast genomes.13-15 While EF-G is essential for general translation, EF4 likely interacts with the bacterial translational machinery in response to certain conditions explaining why its deletion affects bacterial growth or fitness under conditions such as low pH16 or high magnesium (Mg2+) concentrations17 but is a non-essential protein during growth in both rich and poor medium.18-22 In agreement with growth defects, EF4-deficient bacteria exhibited a significantly slower protein translation rate and a delay in overcoming ribosome maturation defects at pH 4 in vivo.16 In addition, EF4 deficiency is known to cause hypersensitivity of E. coli to potassium tellurite and penicillin.19 While the fidelity of translation in vivo does not seem to be affected by the absence of EF4, addition of the purified EF4 has been shown to increase the fraction of active protein synthesized in vitro.18
Nierhaus and coworkers reported that, in vitro, EF4 can catalyze reverse translocation (also called back-translocation), the movement of tRNAs from E and P to P and A sites, respectively.18 Based on the observation that EF4 binds to the POST state with higher affinity than to the PRE state, and that EF4-dependent GTP hydrolysis has a higher turnover rate with the former, it was proposed that the POST complex serves as the substrate of EF4.23 Cryo-EM characterization of the ribosome complex resulting from incubation of the POST complex with EF4, revealed the deacylated tRNA in the classical P site whereas the peptidyl-tRNA was found to occupy a site distinct from classical A/A site (named A/L site, “L” for LepA-induced) with acceptor arm shifted away from the PTC.23 The tRNA was predicted to fall back into the classical A site upon EF4 release from the ribosome. The authors proposed that the resulting complex did not represent a classical PRE complex but rather a back-translocation intermediate state.23 A series of in vitro kinetic assays showed that ribosome back-translocation in the presence of EF4 proceeded through at least 3 intermediate states but also highlighted the relative slowness of the proposed EF4-mediated back-translocation process raising the possibility that POST complex is not the main substrate of EF4.24 Interestingly, single-turnover experiments and single-molecule Förster resonance energy transfer (smFRET) measurements showed that EF4 prefers the PRE state ribosomes and this interaction occurs in a competitive fashion with EF-G,25 which is not surprising given their significant structural similarity discussed below. Therefore, the PRE rather than the POST complex could serve as the substrate and the EF4-mediated PRE complex-like state could arise from its interaction with the PRE complex rather than back-translocating the POST complex.25 Curiously, while Fredrick and co-workers argued against back-translocation26 based on the fact that EF4 failed to promote back-translocation under various conditions including in the mRNA toeprinting assay reported earlier18 and failed to accelerate codon-anticodon complex movement within the 30S subunit of POST complex,24 Qin and co-workers were able to repeat the mRNA toeprinting assay with wild-type and numerous mutant EF4 proteins27. While the biological substrate of EF4 still remains ambiguous, it should be noted that the complexes arising from incubating both the PRE and POST complexes with EF4 are similar and can be rapidly converted to POST complex by EF-G.25 Competition between EF4 and EF-G for the PRE complex has been proposed to transiently slow down polypeptide elongation, thereby facilitating co-translational protein folding.25 Despite comparable affinity for the PRE complex, under normal growth conditions EF-G is about 50-fold more abundant in cells than EF4, suggesting a minor role played by the latter. However, under certain conditions such as high Mg2+ concentration and low pH, EF4 abundance in cytoplasm is believed to increase about 2–3-fold due to its release from membranes, (where it is likely stored under favorable growth conditions17,28), thereby rendering it a more potent competitor for EF-G.
On the other hand, EF4 has been implicated in increasing the elongation rate at above physiological Mg2+ concentrations.17 Although under mild to moderate stress conditions EF4 possibly recruits the stalled ribosomes to resume proper translation, it appears to be harmful during severe stress such as that caused by antimicrobial treatment where deletion of EF4 increased the survival of E. coli after treatment with several antibiotics.29 In this context EF4 was reported to act in a pathway leading to accumulation of reactive oxygen species, thereby facilitating bacterial self-destruction in response to stress-mediated damage to cells.29 This might occur through the reported inhibitory effect of EF4 on the action of transfer-mRNA (tmRNA) in targeting potentially toxic truncated proteins, arising from stress-induced damages to mRNA, to the proteasome.19,29 Insertion of truncated proteins into the cell membrane has been proposed to disturb the respiratory chain leading to the accumulation of reactive oxygen species, and ultimately causing self-destruction.30 Thus, EF4 seems to have 2 functions depending on the severity of stress, it either helps to protect cells by allowing stalled translation to resume at low-to-moderate levels of stress, or leads to bacterial self-destruction at high-levels of stress.29
Interestingly, based on recent ribosome profiling experiments, the loss of EF4 significantly affects the average ribosome density of many mRNAs even in unstressed cells.26 This suggests a function in translation initiation rather than elongation. Fredrick and co-workers propose that EF4 plays a role in ribosome biogenesis and its deficiency could lead to the production of ribosome subunits compromised in initiation stage or, alternatively, EF4 could be directly involved in the initiation process by catalyzing a conformational change in the ribosome affecting interaction with the mRNA Shine-Dalgarno region.26 Taken together, while EF4 seems to aid bacteria in adapting translation to temporary unfavorable conditions as well as affecting translation in general, many aspects of EF4 functioning and importance need further clarification.
BipA
Even more functionally perplexing than EF-G and EF4, is their paralog BipA (Bactericidal/permeability-increasing protein-inducible protein A) that is present in most of the studied bacterial and chloroplast genomes.13,14 BipA has been implicated in regulating a variety of cellular processes including bacterial virulence, symbiosis, resistance to host defenses and antibiotics, swarming motility, biofilm and capsule formation.31-40 As is the case with EF4, BipA is not required under optimal growth conditions but becomes an essential factor for bacterial survival at low temperature, nutrient depletion, and various other stress conditions.32,39,41 Similarity to EF-G and EF4 led to the speculation that BipA affects translation through directly interacting with the ribosome. This is in line with the finding that wild-type (fully modified) ribosomes seem to depend on BipA for the translation of specific mRNAs.42 Given that BipA is able to bind to 70S ribosome in a GTP-dependent manner and its GTPase activity is enhanced in the presence of ribosome as well as inhibited by thiostrepton (characteristic features of trGTPase factors),31,34,43,44 BipA likely functions as an elongation factor that regulates the translation of certain mRNAs under specific stress conditions. Similar to EF4, BipA can inhibit tmRNA tagging, underlining its role as an elongation factor, while its deficiency does not affect the fidelity of translation.19 Curiously, Salmonella enterica BipA has been shown to interact with either 70S ribosome or 30S subunit depending on the relative abundance of GTP and the stress alarmone ppGpp (guanosine-3′, 5′-bis pyrophosphate)31; BipA interacts with 70S ribosome under normal growth conditions but is found interacting with the 30S subunit during amino acid starvation and under sub-optimal growth temperature when the level of ppGpp is increased.31
In addition, a recent study links BipA to ribosome biogenesis as bipA deletion results in perturbed 50S subunit processing and assembly particularly at low temperature.45 Although the evidence for BipA involvement in ribosome biogenesis and/or functioning in translation is mounting, its exact role remains elusive.
Structural comparison of isolated EF-G, EF4, and BipA
Structures of the shared and unique domains of EF-G, EF4, and BipA
While the biological functions of EF-G, EF4, and BipA vary considerably, they all share notable structural similarity.13,43,46-49 All 3 proteins consist of 5 domains, out of which 4 (domains I, II, III, and V) are topologically equivalent (Fig. 1A and 1B). The N-terminal G domain, also named domain I, consists of a central 6-stranded β-sheet surrounded by 5 α-helices (Fig. 1B) and contains the GTP/GDP binding site universally conserved among trGTPase proteins and the homologous Ras superfamily GTPases.15 The G domain contains conserved mobile elements termed switch I, switch II, and the P loop, which are essential for GTPase activation and mediate conformational changes as discussed later. Notably, while the G domains of EF-G, EF4, and BipA are structurally very similar, there is a 95 residue long G' sub-domain insertion is found only in EF-G (Fig. 1C). Domain II contains the signature twisted β–barrel motif shared among translational GTPases (Fig. 1B). Domains III and V contain 4-stranded β-sheets flanked by 2 α-helices on one side (Fig. 1B), a common α/β-motif referred to as the ribonucleoprotein (RNP) or RNA recognition motif (RRM) found in many RNA-binding proteins. While EF4 and BipA lack the region corresponding to EF-G domain IV comprising a unique α/β fold, both have additional C-terminal domains (CTD) that spatially occupy a position between domains III and V (Fig. 1B). The additional CTD is a structural feature observed only in BipA and EF4 trGTPase families.46,48 The CTDs of EF4 and BipA are unique having folds that lack similarity to one another as well as to other known proteins.46,48 The CTD of EF4 comprises one long α-helix cradled by 4 short strands of β-sheet48 (Fig. 1B). The CTD of BipA consists of 2 crossed β-sheets (comprising 2 and 4 β-strands, respectively) wrapped by 3 short α-helices forming a nearly equilateral triangle46,49 (Fig. 1B).
Figure 1.
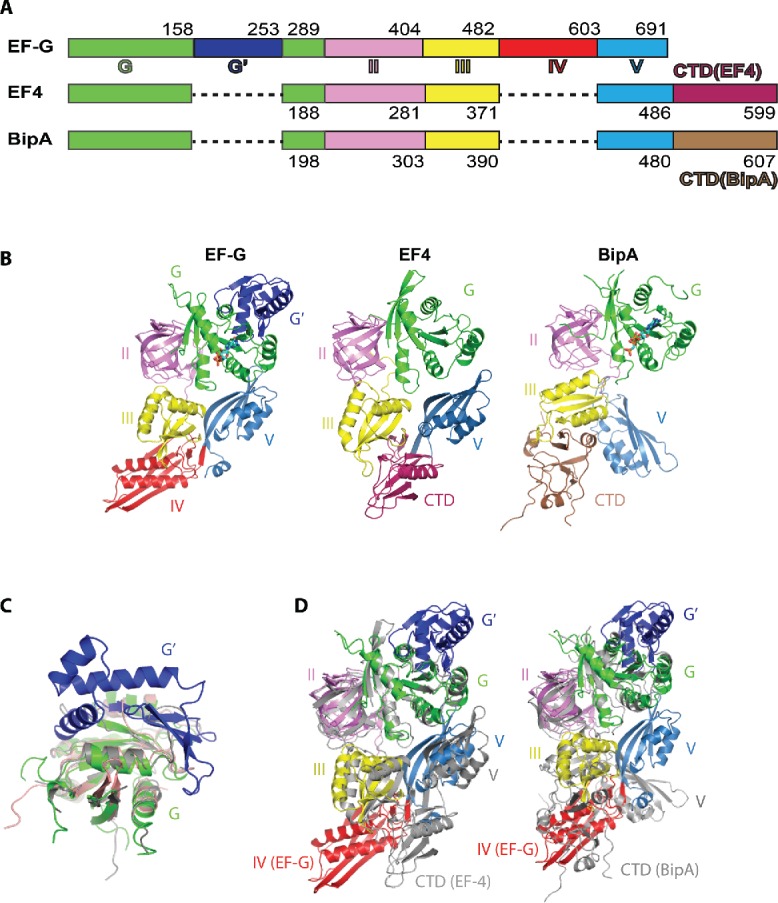
Structural comparison of isolated EF-G, EF4, and BipA. (A) Schematic diagram depicting the domain arrangement of EF-G, EF4, and BipA. (B) Overall structures of isolated EF-G (Protein Data Bank ID: 2BM0), EF4 (PDB ID: 3CB4), and BipA (PDB ID: 5A9W). The same color scheme is used throughout this work unless otherwise stated. Nucleotide bound to the G domain is shown as sticks for EF-G and BipA. EF4 is in apo form. (C) Comparison of domain G of EF-G (colored as in panel B), EF4 (colored gray), and BipA (colored salmon). G' insertion is a characteristic feature of the EF-G protein. (D) Comparison of the domain arrangement of EF-G (colored as in panel B) with EF4 (colored gray on left) and BipA (colored gray on right) by aligning the G domains.
Overall structures of EF-G, EF4, and BipA
Even with 4 out of 5 domains containing similar folds and occupying roughly the same topological position, the extra domains characteristic of EF-G, EF4, and BipA, lead to significant structural variations in the overall conformation of these proteins. First of all, the spatial arrangement of the domains within EF-G, EF4, and BipA proteins is different. Compared with EF-G, the orientation of domain II of both EF4 and BipA with respect to G domain remains largely unchanged, however, the orientation of domain III is rotated in EF4 and BipA (Fig. 1D). While the β-sheet in domain III of EF-G and EF4 has a similar positioning pointing toward domain IV (EF-G) or CTD (EF4), the significant rotation of domain III in BipA results in ∼13 Å movement of the β-sheet toward domain II (Fig. 1D). The most striking difference is observed in domain V, which directly contacts the G domain in EF-G and EF4, but rotates almost by 90° in BipA resulting in over 20 Å distance to G domain (Fig. 1D). While the CTD of BipA occupies a similar position as the domain IV of EF-G, the CTD of EF4 does not (Fig. 1D). Consequently, there is very little spatial overlap between the CTDs of EF4 and BipA. The differences in global conformation due to the distinct domain arrangements of EF-G, EF4, and BipA likely underlie their differing functions in protein synthesis.
Since GTPases are molecular switches known to undergo conformational changes in response to G factor binding and hydrolysis,50-52 it is rather curious that structural comparison of apo forms of EF-G and BipA with non-hydrolysable GTP analog and GDP bound forms reveals very little variation.46,49 This is supported by isothermal titration calorimetry (ITC) analysis of EF-G interaction with GDP and GTP53; and suggests that the structural changes in trGTPases occur only in the presence of ribosome. To date the structure of EF4 has been solved only in the apo form in the absence of ribosome.
An intriguing feature of BipA is that it has been reported to exhibit different modes of ribosome binding whether in complex with alarmone ppGpp or GTP, namely binding to 30S subunits and 70S ribosomes, respectively.31,43 However, in both crystal and solution structures, BipA in complex with ppGpp resembles that of the apo as well as GDPCP- (non-hydrolysable analog of GTP) and GDP-bound BipA complexes.46,49 It should be noted that the binding affinities of BipA and EF-G for GDP and ppGpp are also similar49 underlining the structural conservation of the nucleotide binding sites among trGTPases and suggesting a similar behavior for BipA.
Structure of ribosome-bound EF-G, EF4, and BipA
Development of the ribosome crystal form lacking the L9 protein paved the way for structural studies of GTPase factors bound to the ribosome.54,55 More recently a new approach was developed to crystallize trGTPases on the ribosome by covalently linking them to the N-terminal domain of L947,56 allowing the characterization of more transient interactions. Consequently, structures of EF-G54,56-60 and EF447 bound to the bacterial 70S ribosome have been characterized. In addition, the structure of BipA in complex with the ribosome was reconstructed using recently advanced cryo-EM methods.46 The structures reveal that these proteins occupy a similar position at the interface of the ribosomal subunits known as the factor-binding site. While the structures of isolated EF-G and BipA appear similar regardless of the occupancy of the nucleotide-binding site or the nature of the bound nucleotide, as discussed above, the structures of EF-G, EF4, and BipA in complex with ribosome illustrate dramatic structural rearrangements occurring in these factors as well as in the ribosome.
Changes upon ribosome binding
EF-G
Although biochemical studies have elucidated the overall role of EF-G in translocation, understanding its exact mechanism requires a detailed knowledge of interactions that occur between EF-G, the ribosome, mRNA, and tRNAs throughout the translocation process. As structural studies of EF-G are covered thoroughly in a recently published review,8 a brief summary is given here. Throughout this paper, rRNA residues and helices are numbered according to standard E. coli nomenclature and helices are prefixed by H for 23S rRNA and h for 16S rRNA
Alignment of G domains of the isolated EF-G and ribosome-bound EF-G shows that although domains III and V are shifted markedly, the most striking conformational change occurs in domain IV50-52,57-62 (Fig. 2A). A comparison of the GTP form of EF-G in the PRE state (tRNAs in the A/P and P/E sites)62 with that in the POST state54 reveals that EF-G undergoes a ∼20° rotation around the universally conserved sarcin–ricin loop (SRL) of the 23S rRNA. This rotation results in a movement of the tip of domain IV by 20 Å during the transition from the PRE to the POST state, consistent with the proposed notion that EF-G rotation around SRL allows domain IV of EF-G to avoid a steric clash with the A site tRNA in the PRE state ribosome.62 Moreover, the G domain and domain V of EF-G in the PRE state interact primarily with 50S subunit (universally conserved sarcin-ricin loop and L7/L12 stalk; and the thiostrepton targeted L11-binding region as well as the adjacent H89, respectively) while domains II, III, and IV interact mainly with 30S subunit (h5 and h15 of 16S rRNA, S12, and decoding region, respectively).
Figure 2.
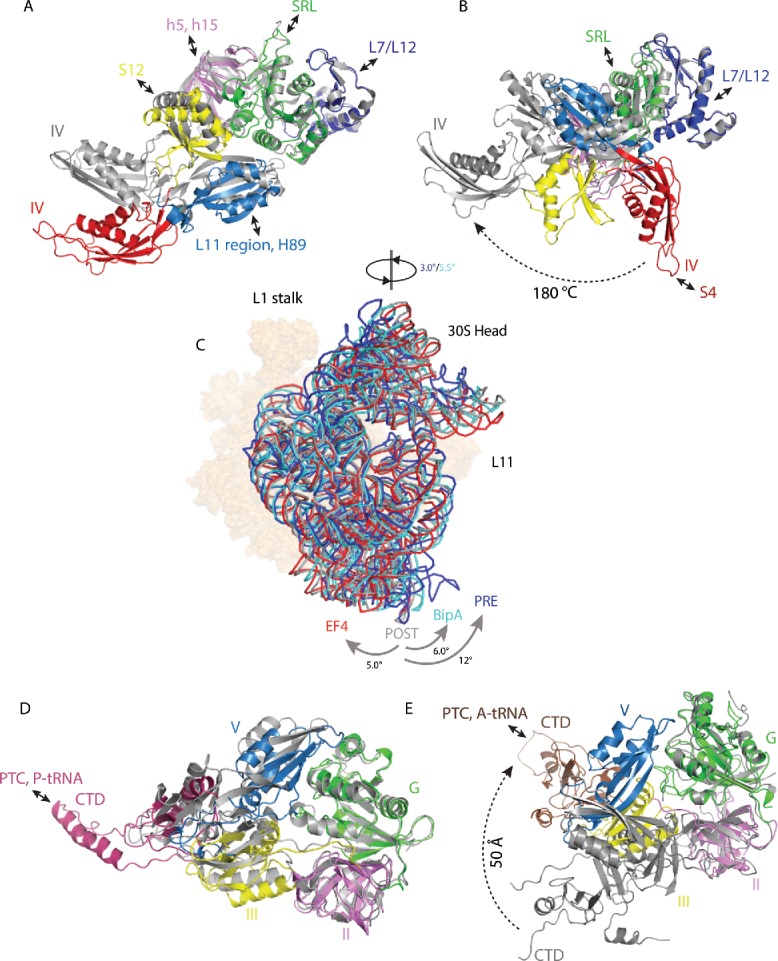
Changes in trGTPase factors and ribosome rotation upon EF-G, EF4, and BipA binding to the ribosome. (A) Comparison of isolated EF-G (PDB ID: 2BM0) with GTP form EF-G in complex with PRE state ribosome trapped by non-hydrolysable GTP analog (PDB ID: 4V90). (B) Comparison of isolated EF-G (PDB ID: 2BM0) with compact form EF-G in complex with PRE state ribosome trapped by non-hydrolysable aminoacyl-tRNA analogs (PDB ID: 4WPO). (C) Comparison of 30S body rotation and head swiveling of POST- (gray) (PDB ID: 4V5F) and PRE- (dark blue) (PDB ID: 5V7C) state ribosomes in complex with EF-G, as well as ribosomes in complex with EF4 (red) (PDB ID: 4W2E) and BipA (light blue) (PDB ID: 5A9Z). For clarity, only 16S rRNA backbone is shown for 30S subunit. 50S subunit is shown as surface in orange. (D) Comparison of isolated EF4 (PDB ID: 3CB4) with ribosome bound EF4 (PDB ID: 4W2E). (E) Comparison of isolated BipA (PDB ID: 5A9W) with ribosome bound BipA (PDB ID: 5A9Z). Ribosome bound trGTPase structures are colored as previously; isolated structures are colored gray. Interactions between trGTPase domains and ribosomal elements are highlighted with double-ended arrows.
Recently, Steitz and coworkers used non-hydrolysable aminoacyl tRNA analogs to prevent deaminoacylation of the P site tRNA and its subsequent translocation into the E site thereby locking the ribosome in PRE state before peptidyl transfer.56 The structure revealed mRNA, tRNAs in the A and P sites, non-rotated ribosome, and domains I and II of EF-G (with bound GDP) in the same conformation as seen in previously determined complexes. Surprisingly, domains III-V adopt a newly observed compact conformation, dramatically different from the elongated one in the PRE complexes,57-59,62 POST complex,54 and in solution regardless of the bound nucleotide.51,61,63,64 In the compact conformation, domains III-IV have rotated with respect to domains I and II, relying on the flexibility of the loop connecting those relatively rigid entities. This results in the flipping of the domain V by ∼180° (compared to isolated EF-G) (Fig. 2B) and a dramatic shift in the positioning of the tip of domain IV pointing it toward S4 protein instead of reaching into the decoding center in the A site as observed in the ribosome-bound elongated EF-G, explaining how domain IV avoids a clash with the A site tRNA before catalyzing translocation. This study also revealed that EF-G adopts the same compact conformation when bound to ribosome trapped in PRE state by antibiotic dityromycin.56 Dityromycin apparently prevents the structural transition from the closed conformation to the elongated one seen in both PRE and POST complexes.56 Overall, the comparison of ribosome-bound EF-G conformations is in line with the hypothesis that the G domain facilitates initial docking of EF-G and the remaining domains move relative to domains G and II during the translocation (upon GTP hydrolysis) in a hinge-like manner.
In addition to conformational changes within EF-G upon ribosome binding, GTPase activation, and translocation, major structural rearrangements take place in the ribosome as well, foremost in the 30S subunit. Compared with the POST state exhibiting an unrotated ribosome,54 the PRE state ribosome62 demonstrates an anti-clockwise rotation of the small subunit by 12° (Fig. 2C). When comparing the crystal structures of PRE complexes reported by different groups, minor differences in the degree of 30S head swiveling and body rotation are observed.58,59 However, cryo-EM analyses of the EF-G–ribosome complex have revealed that the degree of 30S head swiveling and body rotation can vary greatly, by 3°–18° and 4°–9°, respectively.62,65 In addition, the recently described mid-translocation (MID) complex with EF-G and 2 tRNAs (with anticodons between A and P; and P and E sites, respectively) demonstrates 21° head swiveling and 2.7° body rotation compared to classical unrotated ribosome.60 Therefore, the ribosome can adopt various intermediate states of rotation during translocation. Swiveling of the 30S head is necessary to open up a constriction between the platform and the head of 30S allowing the passage of tRNA anticodon stem-loop (ASL) from P to E site.59,60,65,66 Intersubunit rotation and 30S head swiveling are believed to occur in a sequential manner with rotation preceding swiveling, thereby facilitating the directional movement of the mRNA-tRNAs complex.67
EF4
Structural information has demonstrated that EF4 binds to the ribosomes in the same overall orientation as EF-G.23,27,47,68,69 Consistent with the universal trGTPase-binding mode, EF4 forms extensive contacts with both 30S and 50S subunits. The overall conformation of EF4-GDP bound to the ribosome in the crystal structure47 is similar to that of EF4-GDPNP23,27 and EF-4-GDPCP68 in complex with the ribosome in cryo-EM reconstructions. It should be noted, however, that unlike the EF4-GDP-ribosome crystal structure47 and the EF4-GDPCP-ribosome cryo-EM reconstitution68 with tRNAs in classical P site and empty A site, the cryo-EM reconstitutions of the EF4-GDPNP-ribosome complexes reveal 2 tRNAs. In addition to the classical P site, tRNA can also be seen to occupy the A site but in a previously unseen distorted conformation (named A/L and A/4 tRNA in23 and,27 respectively) with acceptor arm shifted away from PTC. A distorted tRNA in the A site is also reported in the very recently published crystal structure of the EF4-GDPCP-ribosome complex69 for which the atomic coordinates are not available yet.
Comparison with the crystal structure of the isolated EF448 (Fig. 2D) reveals that binding to the ribosome affects the positioning of domain V, displacing it by more than 13 Å and rotating by ∼30° so as to avoid a clash with the SRL of the 50S subunit.47 Like EF-G, EF4 contacts both ribosomal subunits. However, the helix-turn-helix (HTH) motif at the tip of the unique CTD occupies the A site (in a position not compatible with the binding of the acceptor stem of a tRNA in classical A site) and reaches into the PTC (Fig. 2D), where it interacts with the acceptor stem of the P site tRNA as well as 23S rRNA in the crystal structure.47 The HTH motif of the CTD in cryo-EM reconstructions is observed to interact with and likely stabilize the acceptor stem of the distorted A site tRNA.27
In contrast to the anti-clockwise rotation of the 30S subunit relative to the 50S subunit observed in ribosome complexes with EF-G, a ∼5° clockwise rotation is seen in the EF4-GDP-ribosome crystal structure (Fig. 2C).47 While a minor ribosome population of the EF4-GDPNP-ribosome complex in cryo-EM analyses also exhibited clockwise rotation, the majority of ribosomes were similar to the unrotated state.27 Interestingly, a cryo-EM analysis of the EF4-GDPCP-ribosome complex revealed, in addition to unrotated ribosomes, a small population of anti-clockwise rotated ribosomes, whereas clockwise-rotated ribosomes were not observed.68 Therefore, the significance of ribosome rotation for EF4 functioning is still unclear and requires further studies.
BipA
Recently, the cryo-EM structure of the ribosome-bound GTP form of BipA was reported unequivocally establishing BipA as a ribosome dependent trGTPase.46 The structure reveals mRNA, A, P, and E site tRNAs, and BipA bound to the same factor binding site in ribosome as EF-G and EF4. The BipA specific CTD occupies the A site of the 50S subunit with the distal loop positioned in close vicinity to the PTC. However, unlike the CTD of EF4 that interacts with the acceptor stem of the tRNA in the P site,47 the CTD of BipA interacts with the A site tRNA. Furthermore, while the overall conformations of isolated and ribosome-bound EF4 are rather similar (Fig. 2D), large conformational changes take place in BipA upon ribosome binding.46 Superimposition of ribosome bound and isolated BipA based on the G domain reveals a significant conformational change for domains III, V, and the CTD (Fig. 2E). The entire domain III makes an anti-clockwise reorientation by over 30° while domain V rotates by almost 90° establishing direct contacts with the G domain. The most striking conformational change, however, is the ∼50 Å rearrangement of the tip of the CTD (Fig. 2E). Similar to the 2 conformations (elongated and compact) revealed for EF-G,56 the structure of isolated BipA (nucleotide-bound or free) exhibits an elongated conformation, and that of ribosome-bound BipA a compact one.
The BipA-ribosome structure with both A and P site tRNAs demonstrates a novel intermediate state of the rotated ribosome. Compared with the structure of the ribosome with EF-G trapped by fusidic acid in an un-rotated POST state,54 a 6˚ anti-clockwise rotation of the 30S body and a 5.5˚ swiveling of the 30S head can be seen46 (Fig. 2C) resembling the ribosome in PRE state in complex with EF-G57 but significantly varying from the clockwise rotated ribosome in complex with EF447 (Fig. 2C).
All in all, when comparing the isolated and ribosome-bound structures of EF-G, EF4, and BipA, it becomes evident that major rearrangements take place in domains III, IV (in case of EF-G), V, and the CTDs (in case of EF4 and BipA), while domains I and II are relatively rigid.
Interaction with the L10–L12 stalk
The L10–L12 stalk, composed of L10 protein and 4–6 copies of L7/L12 as well as its base comprising the L11 region (L11 protein and 23S rRNA helices H43 and H44), is an extremely mobile element of the ribosome and flexibility can be a major obstacle in structural studies. In the majority of the ribosome crystal structures to date, this stalk is almost totally disordered.
It is believed that the dynamic nature of the L10–L12 stalk is essential for its function in “catching” and “handing over” trGTPase factors to the ribosome.1,70 Accordingly, the first contacts of EF-G, EF4, and BipA with the ribosome likely involve the L10–L12 stalk.1 EF‑G was reported to initially dock with the ribosome by contacting the CTD of L12 through its G′ domain.1 However, the affinity of EF-G for isolated L12 is rather low71 and it seems likely that interactions with the SRL of the 50S subunit act to further stabilize the interaction between EF-G and the ribosome. Surprisingly, the structure of the POST complex with EF-G trapped by fusidic acid54 revealed the structure of the L10–L12 stalk in previously unseen detail. The base of the stalk, consisting of L10 protein and 4 copies of the NTD of L12, can be seen at low resolution. The stalk appears to be bent toward EF-G as compared with the structure of the isolated stalk.1,57 Most importantly, the CTD of one of the L12 molecules can be seen in high resolution interacting with the G' subdomain of EF-G (Fig. 3A) as well as with the N-terminal domain (NTD) of L11.54 In the PRE complex with EF-G,57 one copy of the CTD of protein L12 can also be seen interacting with G′ domain of EF-G. However, a structural comparison of the PRE and POST complexes57 reveals a remarkable change in the positioning of the CTD of L12 with respect to EF-G (Fig. 3A), which could be relevant for the release of inorganic phosphate (Pi) following GTP hydrolysis. Indeed, mutations in the CTD of L12 that disrupt its interactions with the G′ domain of EF-G, inhibit Pi release without affecting GTPase activation.72 In addition to initial docking to the ribosome, the interaction between L12 protein and the G' subdomain of EF-G has been shown to be important for the conformational coupling between GTP hydrolysis, release of Pi, and unlocking of the ribosome.63,70 Given that the G' domain is unique to EF-G; its interaction with the CTD of L12 is a characteristic feature of EF-G. The precise role of the G' domain in promoting GTP hydrolysis by EF-G, however, remains to be determined.
Figure 3.
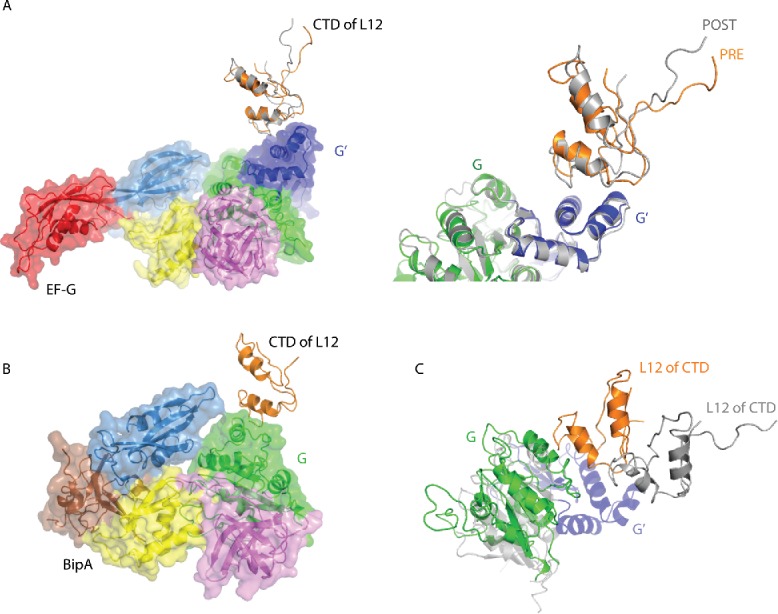
Interaction of the CTD of the L10–L12 stalk protein L12 with EF-G and BipA. (A) Comparison of the CTD of L12 interaction with EF-G in PRE (PDB ID: 4V90) (EF-G colored as previously) and POST (PDB ID: 4V5F) (EF-G colored gray) complexes. The CTD of L12 in PRE complex is shown in orange. Close-up of the EF-G G' domain and the CTD of L12 interaction interface is shown right. (B) CTD of L12 interaction with BipA G domain (PDB ID: 5A9Z). (C) Comparison of CTD of L12 interaction with EF-G in POST complex (PDB ID: 4V5F) and BipA (PDB ID: 5A9Z) by aligning the 23S rRNAs. BipA is colored as previously; POST complex is colored gray with G' domain unique to EF-G highlighted in blue.
While the majority of the L10–L12 stalk was disordered in the structure of ribosome in complex with EF4,47 the complex with BipA clearly shows the entire L11 region as well as one copy of CTD of L12.46 Furthermore, a large density corresponds to the NTD of L12 associated with the long helix in L10 is also visible. The CTD of L12 comes into contact with the universally conserved G domain of BipA (Fig. 3B), unlike the interaction made with the G' domain of EF-G54,57 (Fig. 3A). This newly observed interaction interface involves 2 helices of L12 CTD and one helix of BipA G domain. Considering that the G domain is highly conserved, the interaction between the CTD of L12 and the G domain observed in the BipA-ribosome complex46 could be universal to all trGTPase proteins that lack the G' domain, such as IF2, EF-Tu, RF3, and EF4.
Comparison of the structures of EF-G bound to ribosome in the POST state54 and the BipA-ribosome complex46 reveals a different location of the CTD of L12 protein (Fig. 3C). Namely, the L12 CTD of BipA-ribosome complex would clash with the G' domain of EF-G. Compared with domain V of EF-G, domain V of BipA is located closer to the stalk base, which results in a large conformational change of the L11 protein as well as 23S rRNA helices H43 and H44 in order to avoid a structural clash.
Universal GTPase mechanism
Apart from the G' subdomain insertion in EF-G, the sequences and structures of the G domains of EF-G, EF4, and BipA are highly conserved (Fig. 1C). Also, the G domains of these trGTPases interact with the same region of the ribosome and show little conformational variation upon ribosome binding (Fig. 2A-C). In particular, all trGTPases contain a conserved histidine residue important for efficient GTP hydrolysis on the ribosome.13 The role of His87 (T. thermophilus numbering, His92 in E. coli) in EF-G is played by His81 (E. coli numbering) and His78 (E. coli numbering) in EF447,73 and BipA,46 respectively. The cryo-EM structures of BipA-ribosome46 and EF4-ribosome complexes68 shows the SRL (most likely the A2662 residue) directly contacting the catalytic residues His78 in BipA and His81 in EF4, thereby placing it within interacting distance to the bound GTP analog. This correlates well with the positioning of both His87 in EF-G and His84 in EF-Tu bound to ribosome57-59,74 and underlines the conservation of the GTP hydrolysis mechanism between these proteins.
Based on the recent structures of ribosome bound with EF-G,57-59 the substrate-promoted catalytic mechanism prevails; His87 activates the γ-phosphate, which then abstracts a proton from a water molecule, and the resultant hydroxide ion attacks the γ-phosphate leading to GTP hydrolysis. Thus, the positively charged His87 functions both to initiate the reaction and to stabilize the transition state.57 GTP hydrolysis leads to a series of changes in the switch I, switch II, and P-loop regions, which results in the reorientation of domain IV coupled with 30S swiveling that is believed to promote translocation of the anticodon ends of tRNAs in the ribosome as discussed in next section.
From structure to function
EF-G
The most urgent question is how EF-G binding and GTP hydrolysis alter the structure of EF-G and the ribosome thereby promoting the coupled translocation of mRNA and tRNAs through the ribosome. Based on the high-resolution structures of EF-G in complex with ribosome prior to,56-60 mid- 75, and post-54 translocation, as well as numerous cryo-electron microscopy studies,2,9,62,65,76-78 understanding of how EF-G catalyzes the translocation of the mRNA-tRNAs complex is finally starting to emerge at the atomic level.
In light of the newly characterized compact structure of EF-G on the ribosome, Steitz and co-workers propose that GTP-form EF-G likely engages both the rotated and un-rotated ribosome through a compact structure, thereby avoiding domain IV clashing with the A site tRNA.56 Formation of the rotated state8,79,80 immediately follows binding to the un-rotated ribosome. Following rotation, domain IV is able to extend toward the A site ASL, thereby adopting a conformation seen in the cryo-EM reconstitution of the PRE complex.62 Upon extending toward the A site, domain IV contacts the intersubunit bridge B2a, resulting in the universally conserved nucleotides A1913 of 23S rRNA and A1492 and A1493 of 16S rRNA involved in decoding to adopt different conformations from those seen in other rotated ribosome structures81-83 as well as those observed in the POST complex.54 This transition state is therefore formed specifically upon EF-G binding to promote translocation. The large swiveling of the 30S head observed in the EF-G–ribosome PRE complexes58,60 is proposed to open a ∼20 Å path required for tRNA translocation between the P and E site on the small subunit that is otherwise constricted by 30S head and platform.84
While GTP hydrolysis is known to precede tRNA-mRNA translocation,10,85,86 the exact timing and mechanism of how these processes are linked, needs further clarification. According to the current view, concomitant with EF-G transitioning from compact to elongated form, the conserved histidine is placed in an optimal position relative to SRL for catalyzing GTP hydrolysis. Rapid GTP hydrolysis upon ribosome binding is believed to precede and greatly accelerate the rate-limiting conformational changes resulting in unlocking of the ribosome that is required for translocation followed by re-locking.85
GTP hydrolysis is sensed by the switch I and II regions87 that are disordered in isolated EF-G but are stabilized upon ribosome binding.57-59 The changes in switch I and II are likely communicated to domain III and result in a rearrangement of domains III-V with respect to domains I and II so that the tip of domain IV moves deeper into A site as revealed by the mid-translocation (MID) complex (Fig. 4A).75 The interactions formed between the tip of EF-G domain IV, the ASL, and the corresponding mRNA codon are similar in the MID75 and POST54 translocation complexes, suggesting that they are maintained and likely contribute to correct codon-anticodon pairing during A to P site transitioning, thereby helping to avoid a frameshift during translocation. The importance of the movement of domain IV of EF-G in translocation is corroborated by smFRET results revealing at least 2 different conformations of ribosome-bound EF-G domain IV in solution, corresponding to PRE and POST states.88
Figure 4.
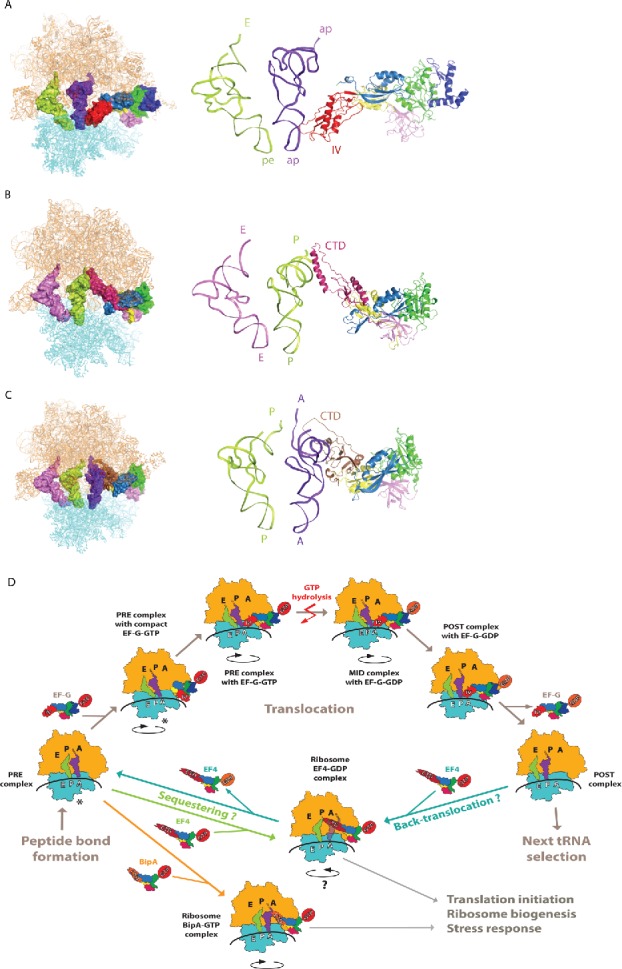
Structural insight into EF-G, EF4, and BipA functioning in translation. (A) Interaction of EF-G domain IV with the ASL of tRNA as revealed by the mid-translocation (MID) complex structure75 shown as surface on the ribosome (left) and as a cartoon in close-up (right). (B) Interaction of EF4 CTD with the P site tRNA in PTC region.47 (C) Interaction of BipA CTD with the A site tRNA in PTC region.46 trGTPases are colored as previously, ribosome 30S and 50S subunits are colored cyan and orange, respectively. tRNAs are colored based on the positioning of their ASL with respect to the decoding center on 30S. (D) Schematic representation of EF-G, EF4, and BipA functioning in translation based on recent structural studies. In short, after peptide bond formation, EF-G recognizes the PRE translocation complex. EF-G first interacts with the ribosome in its compact conformation. Conformational changes in EF-G lead to the transition into elongated form with domain IV extending toward the decoding center in A site coupled to the stabilization of the anti-clockwise rotation of the 30S subunit. Following GTP hydrolysis, the interaction between domain IV of EF-G and the ASL of A site tRNA is likely maintained as the tRNAs-mRNA complex is shifted one codon relative to the ribosome and the 30S subunit returns to the unrotated state. GDP form EF-G dissociates from the POST translocation leaving it ready for next cycle of translation elongation. Asterisk highlights that the PRE complex fluctuates between unrotated and rotated state, both of which are recognized by EF-G and observed in complex with compact EF-G. Under stress conditions, EF4 interacts with either POST “mis-translocation” complex and reverts it back to PRE state allowing EF-G another chance at correct translocation. Alternatively, EF4 as well as BipA can compete with EF-G for the PRE complex and regulate translation in response to stress by likely affecting co-translational protein folding or translation of specific proteins.
Domain IV of EF-G likely serves as a steric block hindering tRNAs from sliding back by occupying the A site as 30S subunit reverts to the un-rotated state upon GTP hydrolysis thereby advancing tRNAs into classical P and E sites observed in the POST complex.54,89 The role of EF-G domain IV in translocation is covered in great detail in recent review.8
Upon GTP hydrolysis and Pi release switch I becomes disordered and EF-G domains II and III move apart.54,63 EF-G relaxing, due to the loss of inter-domain contacts, allows the ribosome to return to the un-rotated state. The GDP form EF-G then dissociates from the ribosome as domain III contacts with 30S and domain V contacts with L11 stalk of the 50S are disrupted.63,90
In brief, ribosome binding and GTP hydrolysis controls the positioning of EF-G domain IV via switch regions and rearrangement of domain III-V with respect to domains I and II. The coordinated action between domain IV and the 30S head swiveling is essential for translocation. Although the process of translocation is intrinsic to the ribosome, EF-G increases its efficiency and biases it in the forward direction. Schematic overview of the translocation process based on available structural and biochemical information is shown in Fig. 4D.
EF4
Despite the fact that EF4 was reported to catalyze back-translocation a decade ago, its precise mechanism remains notoriously elusive. In addition to the shortage of structural evidence, several biochemical studies have called into question the proposed function of EF4 recognizing the POST complex and back-translocating the mRNA-tRNAs complex.25,26
Crystallography47 reveals that the CTD of GDP form of the EF4 reaches into the PTC of the ribosome and contacts the acceptor stem of tRNA in the P site (Fig. 4B) as well as other elements of the PTC. The importance of the CTD of EF4 agrees with the finding that its C-terminal 44 amino acids constituting a flexible sub-domain, while not required for ribosome binding and intrinsic GTPase activity, are important for efficient GTPase activity on the ribosome.73 The clockwise rotation observed in the crystal structure causes the 16S rRNA G530-loop and S12 to shift thereby widening the DC. This conformational change could facilitate the accommodation of the back-translocated tRNA into the A site whereas the translocation of the acceptor stem of tRNA is likely mediated by the CTD of EF4.47 Like EF-G, EF4 may catalyze the unlocking of the ribosome and then bias the Brownian movement of tRNA in the opposite direction. Alternatively, Steitz and coworkers suggest that the CTD of EF4 protects the aminoacylated tRNA in the P site from hydrolysis but in order to do so, the tRNA in the A site would have to be displaced due to steric clashes.47 The widening of DC in response to clockwise rotation could facilitate the release of tRNA by perturbing codon-anticodon interactions in the A site. Therefore, the crystal structure is consistent with EF4 functioning either as a back-translocase or a ribosome sequester (Fig. 4D).47
Recently27 it was proposed that EF4 facilitates back-translocation via its CTD disengaging the tRNA 3′-CCA end from the PTC as well as stabilizing the tRNA in the A/4 site. This is in agreement with previous studies showing that the movement of the 3′-end of peptidyl-tRNA is decoupled from the movement of the rest of the core regions of the ribosome-bound tRNAs, as well as the mRNA, in various steps of the back-translocation process.24 The tRNA remodeling function of EF4 is proposed by Steitz and co-workers based on the new EF4-GDPCP-ribosome crystal structure.69 Namely, the displacement of the CCA end of the tRNA in the A site away from the PTC is functionally significant either by helping to release the deacyl-tRNA from the A site under stress, unlocking a stalled ribosome, or facilitating protein folding.
Taken together, the previously prevailing view on EF4 mediating back-translocation is currently under debate. Structures of EF4-ribosome complexes in various states as well as additional biochemical assays would greatly boost our understanding of EF4 function and its precise mechanism during protein synthesis.
BipA
The cryo-EM reconstruction of the BipA-ribosome-tRNA complex revealed that in addition to interacting with 23S rRNA, the CTD of BipA likely interacts with the A-site tRNA acceptor stem and D-loop region (Fig. 4C).46 This observation is consistent with the C-terminal sequence of BipA being rich in basic residues capable of preferentially binding with nucleic acids, as well as with biochemical data demonstrating a significant role for the C-terminal helix of BipA in ribosome binding.43 Furthermore, the CTD region comprising residues 542-552, which was disordered in the BipA-ribosome reconstitution without tRNA, project deeply into the PTC region surrounded by the 5′ and 3′ ends of the tRNA (Fig. 4C) where the peptide transfer takes place.46 While the EF4-ribosome structure47 shows that the CTD of EF4 also reaches into the PTC, it interacts with the acceptor stem of the P-site tRNA instead (Fig. 4B), as discussed in the previous section. BipA binding leads to anti-clockwise rotation of the ribosome and is compatible with tRNA binding to the A site.46 While both BipA and EF4 confer a growth advantage to bacteria under stress conditions, they appear to accomplish this via different mechanisms likely resulting from the varied location of their CTD in the PTC. In fact, no other trGTPase or stress response factor is known to interact with the A-site tRNA in a similar fashion as BipA hinting at a novel mechanism. However, the precise mechanism of BipA functioning on the ribosome and how it links to its cellular role needs further investigation. Currently, 2 views are prevailing. First, similar to classical trGTPases and EF4, BipA has a regulatory role in protein translation (Fig. 4D).18 The second view is that BipA is a ribosome assembly factor reminiscent of GTPases like Era, EngA, and CgtAE.42,45
BipA binds to the same region of the ribosome as the structurally similar trGTPases, EF-G and EF4 as well as elongation factor Tu (EF-Tu), initiation factor 2 (IF2), and release factor 3 (RF3), all with established roles in translation. In particular, BipA on the ribosome is in active form with the proposed catalytic residue and bound GTP analog positioned close to the SRL of 23S rRNA46 supporting its classification as a bona fide translational factor. On the other hand, when the effect of BipA deletion on ribosome biogenesis was studied, phenotypes often associated with defective ribosome assembly, such as altered subunit ratios and accumulation of 50S precursor particles with partially processed 23S rRNA, were observed.45 This finding suggests that BipA is involved in the production of the 50S subunit. These two proposed functions of BipA may not be mutually exclusive. For example, BipA may be involved in regulating the translation of specific mRNAs whose products act as assembly factors. Indeed, BipA has been reported to be involved in the expression of stress response protein.32,45 In other words, the impairment of 50S assembly may be an indirect result of BipA regulating the translation of specific assembly factors. Curiously, deletion of rluC gene suppresses the ribosome assembly defects of BipA deletion.45 Note that during 50S assembly RluC introduces 3 pseudouridines into 23S rRNA positions (955, 2504, and 2580) close to the PTC in mature ribosome.91,92 This finding supports a link between the functioning of BipA in both translation and ribosome assembly mediated by positioning its CTD close to the PTC and A site tRNA.
The BipA ribosome-binding mode has been reported to differ depending on the cellular levels of GTP and ppGpp.31 Comparison of ppGpp bound BipA with GTP/ribosome bound BipA shows that protrusion of the additional diphosphate moiety at the 3′ hydroxyl of ppGpp results in a steric clash with the SRL46 providing structural insight for the observation that ppGpp-BipA associates with 30S subunits rather than 70S ribosomes.31 GTP and ppGpp are likely 2 alternative physiologically relevant ligands of BipA; when the cells are entering stress but the intra-cellular level of GTP is still high, BipA could be rapidly released so it can bind to the ribosome in its GTP-bound form and regulate the translation of mRNAs of factors involved in 50S assembly. As the stress progresses, the concentration of other adaptive proteins (e.g. RelE/YoeB, YaeJ) would gradually increase, thus inducing ribosome stalling and slowing down translation.93,94 BipA could then interact with the accumulating ppGpp and bind to 30S subunits potentially further inhibiting translation due to assembly defects.31,43 This is in agreement with the findings discussed above, namely that BipA protein is important for 23S rRNA processing and 50S assembly at low temperatures45 and that ribosomes with specific post-transcriptional modifications (introduced by RluC protein) in 23S rRNA depend on BipA for proper assembly.42
trGTPases and antibiotics
The similarities and differences of trGTPases, both in their functioning in the translation process and between different domains of life, are the basis for elucidating the mechanism of numerous families of antibiotics as well as designing new antimicrobial compounds. While structural studies have provided a wealth of information about the functioning of various antibiotics targeting trGTPases and the ribosome, antibiotics themselves have proved to be a useful tool in obtaining a more detailed picture of the processes occurring during translation.
Contribution of antibiotics in elucidating the mechanism of translocation
Several antibiotics, including sparsomycin, streptomycin, paromomycin, hygromycin B, spectinomycin, micrococcin, and thiostrepton, are known to affect various aspects of translocation. The stabilizing effect of viomycin binding to h44 of 16S rRNA and H69 of 23S rRNA on the hybrid state of the ribosome95-97 was utilized for characterizing the PRE complex with EF-G and the 2 hybrid tRNAs using cryo-EM reconstruction62 as well as obtaining the crystal structure of PRE complex with EF-G.58 Viomycin does not directly contact EF-G in the ribosome but is believed to indirectly inhibit its release from the ribosome.62 Viomycin has also been shown to stimulate back-translocation in vitro.98,99
The antibiotic fusidic acid (FA) is known to prevent EF-G release from the ribosome without interfering with GTP hydrolysis and translocation.5,63,79 Similar trapping on the ribosome has been observed for EF-Tu with the antibiotic kirromycin.100 However, unlike the kirromycin interaction with EF-Tu, FA has a very low affinity for isolated EF-G suggesting that FA does not bind to free EF-G, but rather to a specific conformation of EF-G forming on the ribosome. As such, FA had an instrumental role in characterizing GDP/EF-G bound to the POST state ribosomes and the crystal structure revealed the binding site of FA at the interface between the G domain and domain III in vicinity of the GTPase active site54 (Fig. 5A). Based on this POST complex structure, as well as following structural studies of EF-G stabilized on ribosomes in various states utilizing FA,60,75,76,101 it is believed that FA traps EF-G on the ribosome by locking the switch II of EF-G in a conformation similar to the GTP form even after GTP hydrolysis, thereby preventing the transmission of conformational changes to domains III and IV required for EF-G release from the ribosome. FA presumably binds to EF-G in a specific conformation formed transiently during later stages of translocation after tRNAs have at least partially shifted.5,76,102
Figure 5.
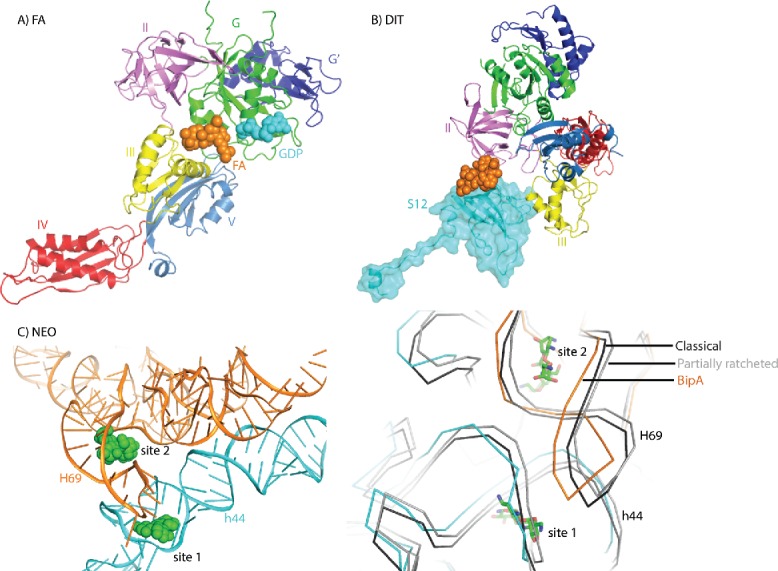
Interplay between ribosome, trGTPases, and antibiotics. (A) Fusidic acid interaction with ribosome bound EF-G (PDB ID: 4V5F). Fusidic acid is shown in orange spheres. GDP bound to EF-G G domain is shown in cyan spheres. (B) Dityromycin interaction with ribosomal protein S12 in the compact EF-G-ribosome structure (PDB ID: 4WQU). Dityromycin is shown in orange spheres and ribosomal protein S12 is highlighted in cyan. (C) Neomycin interaction with ribosome in the BipA-ribosome complex structure (PDB ID: 5A9Z) (left). Neomycin is shown in green spheres. 16S and 23S rRNA are shown in cyan and orange, respectively. Comparison of the neomycin binding sites in the rotated ribosome in complex with BipA (PDB ID: 5A9Z) (same coloring), unrotated ribosome (black) in complex with RRF (PDB ID: 4V54) and partially rotated (gray) ribosome (PDB ID: 4V9C) (right). For clarity, only rRNA backbone is shown. Neomycin is shown in green sticks.
Sordarin was thought to act in a similar fashion as FA on yeast eEF2 (an homolog of EF-G) by also preventing large-scale conformational changes in the trGTPase required for its release after GTP hydrolysis.103 However, unlike FA, sordarin can bind to free eEF2 and form interactions with domains III, IV, and V, but not the G domain, causing substantial conformational changes in eEF2 compared to its apo form as well as isolated and ribosome-bound EF-G.104 Sordarin likely prevents domain III from moving away from SRL, thereby preventing the dissociation of eEF2 from the ribosome.105,106
The antibiotic dityromycin has also been shown to block EF-G associated translocation.107 The crystal structure of dityromycin in complex with the ribosome reveals that it binds to ribosomal protein S12 and would clash with domain III of EF-G in its elongated form108 (Fig. 5B). Unexpectedly, the crystal structure of EF-G trapped to the PRE state ribosome by dityromycin reveals a previously unseen compact conformation of EF-G (Fig. 5B) and suggests that dityromycin binding to protein S12 inhibits translocation by blocking the transition of EF-G into the elongated form with domain IV protruding into the A site.56
Neomycin interaction with the ribosome
Structural and smFRET studies have revealed that aminoglycoside neomycin blocks aminoacyl-tRNA selection, translocation, and ribosome recycling by binding to H69 of 50S subunit 23S rRNA.109-111 While studies of neomycin have underlined the importance of intersubunit rotation in translocation, its major effect on stabilizing an intermediate state of the ribosome can be utilized to enhance trGTPase binding. Indeed, neomycin was used to trap the mid-translocation state ribosome with EF-G and.75 More recently, neomycin was found to greatly enhance the otherwise transient binding of BipA to the ribosome, allowing Gao and coworkers to reconstruct the cryo-EM structure of BipA bound to the rotated ribosome.46
Neomycin was observed to bind to 2 sites, one in h44 of 16S rRNA in close proximity to the tip of H69 (site 1), and another in the major groove of H69 of 23S rRNA at the base of its stem (site 2) (Fig. 5C). These two sites overlap with that in the structures of RRF bound to unrotated110 and partially rotated ribosomes.111 Comparison of these structures with that of ribosome-BipA reveals that, while a minor conformational change occurs in the 16S rRNA surrounding site 1, the tip of H69 of 23S rRNA undergoes a large shift upon 30S rotation leading to its involvement in forming the binding site 1 for neomycin (Fig. 5C). As for site 2, both 16S and 23S rRNA surrounding neomycin move notably, demonstrating a more dynamic feature (Fig. 5C). Curiously, the structure of the mid-translocation ribosome with EF-G reports more neomycin-binding sites, whereas neomycin is not observed in site 2.75 Structural comparison appears to be consistent with the notion that neomycin preferentially binds to H69 when the ribosome adopts the rotated state.110 The neomycin-binding site is too far away to establish any direct interaction with BipA. Thus, neomycin-induced (or –assisted) stabilization of the ribosome configuration likely enhances the binding of BipA to the ribosome. Taken together, neomycin interactions with the ribosome appear to be complex and vary depending on the different functional states of the ribosome as well as the presence of diverse trGTPases.
Conclusion
Thanks to the advances in cryo-EM technology as well as the utilization of various antibiotics in order to trap more transient complexes for structural characterization, the last couple of years have provided invaluable insight into the enigmatic translocation process. Furthermore, the characterization of the interaction of proteins EF4 and BipA with the ribosome provides the structural basis for elucidating how translation could be regulated under certain conditions.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank A.W.H. Khong for help with figures and C. Jelinska for proofreading the manuscript.
Funding
This work was supported by the Singapore National Research Foundation NRF-RF2009-RF001-267 (Y.G.G.) and a Tier II grant from the Ministry of Education (MOE) of Singapore (Y.G.G.).
References
- 1.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 2005; 121:991-1004; PMID:15989950; http://dx.doi.org/ 10.1016/j.cell.2005.04.015 [DOI] [PubMed] [Google Scholar]
- 2.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000; 406:319-22; PMID:10917535; http://dx.doi.org/2682263 10.1038/35018597 [DOI] [PubMed] [Google Scholar]
- 3.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature 1989; 342:142-8; PMID:2682263; http://dx.doi.org/ 10.1038/342142a0 [DOI] [PubMed] [Google Scholar]
- 4.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 2007; 370:530-40; PMID:17512008; http://dx.doi.org/ 10.1016/j.jmb.2007.04.042 [DOI] [PubMed] [Google Scholar]
- 5.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 2009; 15:772-80; PMID:19324963; http://dx.doi.org/ 10.1261/rna.1592509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans 2011; 39:658-62; PMID:21428957; http://dx.doi.org/ 10.1042/BST0390658 [DOI] [PubMed] [Google Scholar]
- 7.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 2010; 466:329-33; PMID:20631791; http://dx.doi.org/ 10.1038/nature09206 [DOI] [PubMed] [Google Scholar]
- 8.Holtkamp W, Wintermeyer W, Rodnina MV. Synchronous tRNA movements during translocation on the ribosome are orchestrated by elongation factor G and GTP hydrolysis. Bioessays 2014; 36:908-18; PMID:25118068; http://dx.doi.org/ 10.1002/bies.201400076 [DOI] [PubMed] [Google Scholar]
- 9.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al.. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell 2007; 25:751-64; PMID:17349960; http://dx.doi.org/ 10.1016/j.molcel.2007.01.027 [DOI] [PubMed] [Google Scholar]
- 10.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 1997; 385:37-41; PMID:8985244; http://dx.doi.org/ 10.1038/385037a0 [DOI] [PubMed] [Google Scholar]
- 11.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor g to translocation and factor recycling on the ribosome. Biochemistry 2002; 41:12806-12; PMID:12379123; http://dx.doi.org/ 10.1021/bi0264871 [DOI] [PubMed] [Google Scholar]
- 12.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell 2002; 108:557-72; PMID:11909526; http://dx.doi.org/ 10.1016/S0092-8674(02)00619-0 [DOI] [PubMed] [Google Scholar]
- 13.Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics 2007; 8:15; PMID:17214893; http://dx.doi.org/ 10.1186/1471-2164-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 2002; 317:41-72; PMID:11916378; http://dx.doi.org/ 10.1006/jmbi.2001.5378 [DOI] [PubMed] [Google Scholar]
- 15.Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr Opin Microbiol 2003; 6:135-9; PMID:12732302; http://dx.doi.org/ 10.1016/S1369-5274(03)00037-7 [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Li Z, Hao J, Qin Y. EF4 knockout E. coli cells exhibit lower levels of cellular biosynthesis under acidic stress. Protein Cell 2014; 5:563-7; PMID:24706296; http://dx.doi.org/ 10.1007/s13238-014-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci U S A 2011; 108:3199-203; PMID:21300907; http://dx.doi.org/ 10.1073/pnas.1012994108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 2006; 127:721-33; PMID:17110332; http://dx.doi.org/ 10.1016/j.cell.2006.09.037 [DOI] [PubMed] [Google Scholar]
- 19.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 2010; 92:157-63; PMID:19925844; http://dx.doi.org/ 10.1016/j.biochi.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dibb NJ, Wolfe PB. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol 1986; 166:83-7; PMID:3514582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colca JR, McDonald WG, Waldon DJ, Thomasco LM, Gadwood RC, Lund ET, Cavey GS, Mathews WR, Adams LD, Cecil ET, et al.. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J Biol Chem 2003; 278:21972-9; PMID:12690106; http://dx.doi.org/ 10.1074/jbc.M302109200 [DOI] [PubMed] [Google Scholar]
- 22.Badu-Nkansah A, Sello JK. Deletion of the elongation factor 4 gene (lepA) in Streptomyces coelicolor enhances the production of the calcium-dependent antibiotic. FEMS Microbiol Lett 2010; 311:147-51; PMID:20735483; http://dx.doi.org/ 10.1111/j.1574-6968.2010.02083.x [DOI] [PubMed] [Google Scholar]
- 23.Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol 2008; 15(9):910-5; PMID:19172743; http://dx.doi.org/ 10.1038/nsmb.1469 [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J Mol Biol 2010; 396:1043-52; PMID:20045415; http://dx.doi.org/ 10.1016/j.jmb.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Chen C, Zhang H, Kaur J, Goldman YE, Cooperman BS. The conserved protein EF4 (LepA) modulates the elongation cycle of protein synthesis. Proc Natl Acad Sci U S A 2011; 108:16223-8; PMID:21930951; http://dx.doi.org/ 10.1073/pnas.1103820108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res 2014; 42:13370-83; PMID:25378333; http://dx.doi.org/ 10.1093/nar/gku1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Yan K, Liu G, Song G, Luo J, Shi Y, Cheng E, Wu S, Jiang T, Lou J, Gao N, Qin Y. EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome. Nat Struct Mol Biol 2016; 23(2):125-31; PMID:26809121; http://dx.doi.org/24362468 10.1038/nsmb.3160 [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto H, Qin Y, Achenbach J, Li C, Kijek J, Spahn CM, Nierhaus KH. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol 2014; 12:89-100; PMID:24362468; http://dx.doi.org/ 10.1038/nrmicro3176 [DOI] [PubMed] [Google Scholar]
- 29.Li L, Hong Y, Luan G, Mosel M, Malik M, Drlica K, Zhao X. Ribosomal elongation factor 4 promotes cell death associated with lethal stress. MBio 2014; 5:e01708; PMID:25491353; http://dx.doi.org/ 10.1128/mBio.01708-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 2008; 135:679-90; PMID:19013277; http://dx.doi.org/ 10.1016/j.cell.2008.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.deLivron MA, Robinson VL. Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J Bacteriol 2008; 190:5944-52; PMID:18621905; http://dx.doi.org/ 10.1128/JB.00763-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss E, Huguet T, Poinsot V, Batut J. The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Mol Plant Microbe Interact 2004; 17:235-44; PMID:15000390; http://dx.doi.org/ 10.1094/MPMI.2004.17.3.235 [DOI] [PubMed] [Google Scholar]
- 33.Qi SY, Li Y, Szyroki A, Giles IG, Moir A, O'Connor CD. Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol Microbiol 1995; 17:523-31; PMID:8559071; http://dx.doi.org/ 10.1111/j.1365-2958.1995.mmi_17030523.x [DOI] [PubMed] [Google Scholar]
- 34.Farris M, Grant A, Richardson TB, O'Connor CD. BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol Microbiol 1998; 28:265-79; PMID:9622352; http://dx.doi.org/ 10.1046/j.1365-2958.1998.00793.x [DOI] [PubMed] [Google Scholar]
- 35.Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O'Connor CD. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol Microbiol 2003; 48:507-21; PMID:12675808; http://dx.doi.org/ 10.1046/j.1365-2958.2003.t01-1-03447.x [DOI] [PubMed] [Google Scholar]
- 36.Overhage J, Lewenza S, Marr AK, Hancock RE. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 2007; 189:2164-9; PMID:17158671; http://dx.doi.org/ 10.1128/JB.01623-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J Bacteriol 2000; 182:2741-5; PMID:10781541; http://dx.doi.org/ 10.1128/JB.182.10.2741-2745.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neidig A, Yeung AT, Rosay T, Tettmann B, Strempel N, Rueger M, Lesouhaitier O, Overhage J. TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol 2013; 13:77; PMID:23570569; http://dx.doi.org/ 10.1186/1471-2180-13-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfennig PL, Flower AM. BipA is required for growth of Escherichia coi K12 at low temperature. Mol Genet Genomics 2001; 266:313-7; PMID:11683274; http://dx.doi.org/ 10.1007/s004380100559 [DOI] [PubMed] [Google Scholar]
- 40.Duo M, Hou S, Ren D. Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl Microbiol Biotechnol 2008; 81:731-41; PMID:18807027; http://dx.doi.org/ 10.1007/s00253-008-1709-6 [DOI] [PubMed] [Google Scholar]
- 41.Eymann C, Homuth G, Scharf C, Hecker M. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol 2002; 184:2500-20; PMID:11948165; http://dx.doi.org/ 10.1128/JB.184.9.2500-2520.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan K, Flower AM. Suppression of DeltabipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J Bacteriol 2008; 190:7675-83; PMID:18820021; http://dx.doi.org/ 10.1128/JB.00835-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.deLivron MA, Makanji HS, Lane MC, Robinson VL. A novel domain in translational GTPase BipA mediates interaction with the 70S ribosome and influences GTP hydrolysis. Biochemistry 2009; 48:10533-41; PMID:19803466; http://dx.doi.org/ 10.1021/bi901026z [DOI] [PubMed] [Google Scholar]
- 44.Mikolajka A, Liu H, Chen Y, Starosta AL, Márquez V, Ivanova M, Cooperman BS, Wilson DN. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol 2011; 18:589-600; PMID:21609840; http://dx.doi.org/ 10.1016/j.chembiol.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhury P, Flower AM. Efficient Assembly of Ribosomes is Inhibited by Deletion of bipA in Escherichia coli. J Bacteriol 2015; 197(10):1819-27; PMID:25777676; http://dx.doi.org/ 10.1128/JB.00023-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar V, Chen Y, Ero R, Ahmed T, Tan J, Li Z, Wong AS, Bhushan S, Gao YG. Structure of BipA in GTP form bound to the ratcheted ribosome. Proc Natl Acad Sci U S A 2015; 112:10944-9; PMID:26283392; http://dx.doi.org/ 10.1073/pnas.1513216112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagnon MG, Lin J, Bulkley D, Steitz TA. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science 2014; 345:684-7; PMID:25104389; http://dx.doi.org/ 10.1126/science.1253525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci U S A 2008; 105:4673-8; PMID:18362332; http://dx.doi.org/ 10.1073/pnas.0801308105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan H, Hahm J, Diggs S, Perry JJ, Blaha G. Structural and Functional Analysis of BipA, a Regulator of Virulence in enteropathogenic Escherichia coli. J Biol Chem 2015; 290(34):20856-64; PMID:26163516; http://dx.doi.org/8070397 10.1074/jbc.M115.659136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aevarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. Embo J 1994; 13:3669-77; PMID:8070397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett 2005; 579:4492-7; PMID:16083884; http://dx.doi.org/ 10.1016/j.febslet.2005.07.016 [DOI] [PubMed] [Google Scholar]
- 52.al-Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure 1996; 4:555-65; PMID:8736554; http://dx.doi.org/ 10.1016/S0969-2126(96)00061-5 [DOI] [PubMed] [Google Scholar]
- 53.Hauryliuk V, Mitkevich VA, Eliseeva NA, Petrushanko IY, Ehrenberg M, Makarov AA. The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc Natl Acad Sci U S A 2008; 105:15678-83; PMID:18836081; http://dx.doi.org/ 10.1073/pnas.0807912105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009; 326:694-9; PMID:19833919; http://dx.doi.org/ 10.1126/science.1179709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selmer M, Gao YG, Weixlbaumer A, Ramakrishnan V. Ribosome engineering to promote new crystal forms. Acta Crystallogr D Biol Crystallogr 2012; 68:578-83; PMID:22525755; http://dx.doi.org/ 10.1107/S0907444912006348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 2015; 160:219-27; PMID:25594181; http://dx.doi.org/ 10.1016/j.cell.2014.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol 2013; 20:1077-84; PMID:23912278; http://dx.doi.org/ 10.1038/nsmb.2645 [DOI] [PubMed] [Google Scholar]
- 58.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science 2013; 340:1235970; PMID:23812721; http://dx.doi.org/ 10.1126/science.1235970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 2013; 340:1235490; PMID:23812720; http://dx.doi.org/ 10.1126/science.1235490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 2013; 340:1236086; PMID:23812722; http://dx.doi.org/ 10.1126/science.1236086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. Embo J 1994; 13:3661-8; PMID:8070396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A 2013; 110:20994-9; PMID:24324137; http://dx.doi.org/ 10.1073/pnas.1311423110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J 2009; 28:2053-65; PMID:19536129; http://dx.doi.org/ 10.1038/emboj.2009.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czworkowski J, Moore PB. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry 1997; 36:10327-34; PMID:9254632; http://dx.doi.org/ 10.1021/bi970610k [DOI] [PubMed] [Google Scholar]
- 65.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al.. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 2010; 468:713-6; PMID:21124459; http://dx.doi.org/ 10.1038/nature09547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science 2009; 325:1014-7; PMID:19696352; http://dx.doi.org/ 10.1126/science.1175275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A 2012; 109:20391-4; PMID:23188795; http://dx.doi.org/ 10.1073/pnas.1218999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar, V, Ero, R, Ahmed, T, Goh, KJ, Zhan, Y, Bhushan, S, Gao, YG. Structure of the GTP form of elongation factor 4 (EF4) bound to the ribosome. J Biol Chem 2016; 291:12943-50; PMID:27137929; http://dx.doi.org/27092003 10.1074/jbc.M116.725945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gagnon MG, Lin J, Steitz TA. Elongation factor 4 remodels the A-site tRNA on the ribosome. Proc Natl Acad Sci U S A 2016; 113:4994-9; PMID:27092003; http://dx.doi.org/ 10.1073/pnas.1522932113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nechifor R, Wilson KS. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J Mol Biol 2007; 368:1412-25; PMID:17395204; http://dx.doi.org/ 10.1016/j.jmb.2007.03.009 [DOI] [PubMed] [Google Scholar]
- 71.Savelsbergh A, Mohr D, Wilden B, Wintermeyer W, Rodnina MV. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J Biol Chem 2000; 275:890-4; PMID:10625623; http://dx.doi.org/ 10.1074/jbc.275.2.890 [DOI] [PubMed] [Google Scholar]
- 72.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J 2005; 24:4316-23; PMID:16292341; http://dx.doi.org/ 10.1038/sj.emboj.7600884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Laurentiis EI, Wieden HJ. Identification of two structural elements important for ribosome-dependent GTPase activity of elongation factor 4 (EF4/LepA). Sci Rep 2015; 5:8573; PMID:25712150; http://dx.doi.org/ 10.1038/srep08573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 2010; 330:835-8; PMID:21051640; http://dx.doi.org/ 10.1126/science.1194460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 2014; 345:1188-91; PMID:25190797; http://dx.doi.org/ 10.1126/science.1255030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci U S A 2013; 110:20964-9; PMID:24324168; http://dx.doi.org/ 10.1073/pnas.1320387110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson KS, Nechifor R. Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J Mol Biol 2004; 337:15-30; PMID:15001349; http://dx.doi.org/ 10.1016/j.jmb.2004.01.013 [DOI] [PubMed] [Google Scholar]
- 78.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 2000; 100:301-9; PMID:10676812; http://dx.doi.org/ 10.1016/S0092-8674(00)80666-2 [DOI] [PubMed] [Google Scholar]
- 79.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 2007; 13:1473-82; PMID:17630323; http://dx.doi.org/ 10.1261/rna.601507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 2011; 18:457-62; PMID:21399643; http://dx.doi.org/ 10.1038/nsmb.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 2012; 18:230-40; PMID:22187675; http://dx.doi.org/ 10.1261/rna.031187.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A 2011; 108:15798-803; PMID:21903932; http://dx.doi.org/ 10.1073/pnas.1112185108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 2011; 332:981-4; PMID:21596992; http://dx.doi.org/ 10.1126/science.1202692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science 2005; 310:827-34; PMID:16272117; http://dx.doi.org/ 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- 85.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 2003; 11:1517-23; PMID:12820965; http://dx.doi.org/ 10.1016/S1097-2765(03)00230-2 [DOI] [PubMed] [Google Scholar]
- 86.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 2007; 25:519-29; PMID:17317625; http://dx.doi.org/ 10.1016/j.molcel.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299-304; PMID:11701921; http://dx.doi.org/ 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- 88.Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci U S A 2014; 111:15060-5; PMID:25288752; http://dx.doi.org/ 10.1073/pnas.1410873111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol 2000; 300:951-61; PMID:10891280; http://dx.doi.org/ 10.1006/jmbi.2000.3886 [DOI] [PubMed] [Google Scholar]
- 90.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci U S A 2006; 103:13670-5; PMID:16940356; http://dx.doi.org/ 10.1073/pnas.0606099103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conrad J, Sun D, Englund N, Ofengand J. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA. J Biol Chem 1998; 273:18562-6; PMID:9660827; http://dx.doi.org/ 10.1074/jbc.273.29.18562 [DOI] [PubMed] [Google Scholar]
- 92.Siibak T, Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA 2010; 16:2023-32; PMID:20719918; http://dx.doi.org/ 10.1261/rna.2160010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 2009; 139:1084-95; PMID:20005802; http://dx.doi.org/ 10.1016/j.cell.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng S, Chen Y, Kamada K, Wang H, Tang K, Wang M, Gao YG. YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res 2013; 41:9549-56; PMID:23945936; http://dx.doi.org/ 10.1093/nar/gkt742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol 2007; 14:493-7; PMID:17515906; http://dx.doi.org/ 10.1038/nsmb1243 [DOI] [PubMed] [Google Scholar]
- 96.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol 2004; 343:1183-94; PMID:15491605; http://dx.doi.org/ 10.1016/j.jmb.2004.08.097 [DOI] [PubMed] [Google Scholar]
- 97.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol 2010; 17:289-93; PMID:20154709; http://dx.doi.org/ 10.1038/nsmb.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell 2006; 24:931-42; PMID:17189194; http://dx.doi.org/ 10.1016/j.molcel.2006.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol 2007; 14:318-24; PMID:17369838; http://dx.doi.org/ 10.1038/nsmb1221 [DOI] [PubMed] [Google Scholar]
- 100.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009; 326:688-94; PMID:19833920; http://dx.doi.org/ 10.1126/science.1179700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng S, Chen Y, Gao YG. Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. PLoS One 2013; 8:e58829; http://dx.doi.org/ 10.1371/journal.pone.0058829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry 2006; 45:2504-14; PMID:16489743; http://dx.doi.org/ 10.1021/bi0516677 [DOI] [PubMed] [Google Scholar]
- 103.Søe R, Mosley RT, Justice M, Nielsen-Kahn J, Shastry M, Merrill AR, Andersen GR. Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2. J Biol Chem 2007; 282:657-66; PMID:17082187; http://dx.doi.org/ 10.1074/jbc.M607830200 [DOI] [PubMed] [Google Scholar]
- 104.Jørgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 2003; 10:379-85; PMID:12692531; http://dx.doi.org/ 10.1038/nsb923 [DOI] [PubMed] [Google Scholar]
- 105.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 2004; 23:1008-19; PMID:14976550; http://dx.doi.org/ 10.1038/sj.emboj.7600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chakraborty B, Mukherjee R, Sengupta J. Structural insights into the mechanism of translational inhibition by the fungicide sordarin. J Comput Aided Mol Des 2013; 27:173-84; PMID:23397219; http://dx.doi.org/ 10.1007/s10822-013-9636-8 [DOI] [PubMed] [Google Scholar]
- 107.Brandi L, Maffioli S, Donadio S, Quaglia F, Sette M, Milon P, Gualerzi CO, Fabbretti A. Structural and functional characterization of the bacterial translocation inhibitor GE82832. FEBS Lett 2012; 586:3373-8; PMID:22841550; http://dx.doi.org/ 10.1016/j.febslet.2012.07.040 [DOI] [PubMed] [Google Scholar]
- 108.Bulkley D, Brandi L, Polikanov YS, Fabbretti A, O'Connor M, Gualerzi CO, Steitz TA. The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation. Cell Rep 2014; 6:357-65; PMID:24412368; http://dx.doi.org/ 10.1016/j.celrep.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Frank J, Gonzalez RLJ. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 2010; 79:381-412; PMID:20235828; http://dx.doi.org/ 10.1146/annurev-biochem-060408-173330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 2007; 14:727-32; PMID:17660832; http://dx.doi.org/ 10.1038/nsmb1271 [DOI] [PubMed] [Google Scholar]
- 111.Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JH, Blanchard SC. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol 2012; 19:957-63; PMID:22902368; http://dx.doi.org/ 10.1038/nsmb.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]


