Abstract
We performed a scan for genetic variants associated with multiple phenotypes by comparing large genome-wide association studies (GWAS) of 42 traits or diseases. We identified 341 loci (at an FDR of 10%) associated with multiple traits. Several loci are associated with a large number of phenotypes; for example, a nonsynonymous variant in the zinc transporter SLC39A8 influences seven of these traits, including risk of schizophrenia (rs13107325: log-odds ratio = 0.15, P = 2 × 10−12) and Parkinson's disease (log-odds ratio = −0.15, P = 1.6 × 10−7), among others. Second, we used these loci to identify traits that share multiple genetic causes in common. For example, variants that increase risk of schizophrenia also tend to increase risk of inflammatory bowel disease. Finally, we developed a method to identify pairs of traits that show evidence of a causal relationship. For example, we show evidence that increased BMI causally increases triglyceride levels.
Introduction
The observation that a genetic variant affects multiple phenotypes (a phenomenon often called “pleiotropy” 1-3, though we will not use this term) is informative in a number of applications. One such application is to learn about the molecular function of a gene. For example, men with cystic fibrosis (primarily known as a lung disease) are often infertile due to congenital absence of the vas deferens; this is evidence of a shared role for the CFTR protein in lung function and the development of reproductive organs 4. Another application is to learn about the causal relationships between traits. For example, individuals with congenital hypercholesterolemia also have elevated risk of heart disease 5; this is now interpreted as evidence that changes in lipid levels causally influence heart disease risk 6.
In these two applications, the same observation–that a genetic variant influences two traits–is interpreted in fundamentally different ways depending on known aspects of biology. In the first case, a genetic variant influences the two phenotypes through independent physiological mechanisms (graphically: P1 ← G → P2, if G represents the genotype, P1 the first phenotype, P2 the second phenotype, and the arrows represent causal relationships7), while in the second case, G → P1 → P2. In some situations, knowing which interpretation of the observation to prefer is simple: for example, it seems difficult to imagine how the reproductive and lung phenotypes of a CFTR mutation could be related in a causal chain. In other situations, interpretation is considerably more challenging. For example, the causal connections between various lipid phenotypes and heart disease have been debated for decades (e.g. 8).
As the number of reliable associations between genetic variants and various phenotypes has grown over the last decade 9, these issues have received increasing attention. A number of recent studies have identified genetic variants associated with multiple traits 10-20; in general, these associations are interpreted as most plausibly due to independent effects of a genetic variant on different aspects of physiology. For example, a genetic variant in LGR4 is associated with bone mineral density (BMD), age at menarche, and risk of gallbladder cancer 16, presumably due to effects mediated through different tissues.
There has also been increasing interest in the alternative, causal framework for interpreting genetic variants that influence multiple phenotypes, which has been formalized under the name “Mendelian randomization” 21-23. Mendelian randomization has been used to provide evidence for (or against) a causal role for various clinical variables in disease etiology 24-30. For example, genetic variants associated with body mass index (BMI) are also associated with type 2 diabetes 27; this is consistent with a causal role for weight gain in the etiology of diabetes.
To date, most studies of multiple traits have been performed genome-wide on groups of traits already known or hypothesized to be related 10;31-33, or via testing small sets of variants for effects on a wide range of traits 20;34. We aimed to systematically perform a genome-wide search for genetic variants that influence pairs of traits, and then to interpret these associations in the light of the causal and non-causal models described above. In this paper, we describe the results of such a search using large genome-wide association studies of 42 traits.
Results
We assembled summary statistics from 43 genome-wide association studies of 42 traits or diseases performed in individuals of European descent (Table 1; two of these GWAS are for age at menarche). These studies span a wide range of phenotypes, from anthropometric traits (e.g. height, BMI, nose size) to neurological disease (e.g. Alzheimer's disease, Parkinson's disease) to susceptibility to infection (e.g. childhood ear infections, tonsillectomy). 17 of these GWAS were performed by the personal genomics company 23andMe, and have not previously been reported (for details of these studies, see Supplementary Data Sets 1-17). For studies that were not done using imputation to all variants in phase 1 of the 1000 Genomes Project 35, we performed imputation at the level of summary statistics using ImpG v1.0 36. We estimated the approximate number of independent associated variants (at a false discovery rate of 10%) in each study using fgwas v.0.3.6 37. The number of associations ranged from around five (for age at voice drop in men) to over 500 (for height).
Table 1. Phenotypes used in this study.
For each study, we show the name of the phenotype, the abbreviation that will be used throughout this paper, the data source, the number of independent autosomal loci identified at a false discovery rate of 10%, and the number of participants in the study. For studies where the data source is 23andMe, a complete description of the GWAS is presented in the Supplementary Material.
| Phenotype | Abbreviation | Data source | Approx # of loci | Approx # of participants, in thousands (cases/controls, if applicable) |
|---|---|---|---|---|
| Neurological phenotypes | ||||
| Alzheimer's disease | AD | 75 | 11 | 17 / 37 |
| Migraine | MIGR | 23andMe | 37 | 53 / 231 |
| Parkinson's disease | PD | 23andMe | 43 | 10 / 325 |
| Photic sneeze reflex | PS | 23andMe | 66 | 32 / 67 |
| Schizophrenia | SCZ | 59 | 222 | 34 / 46 |
|
Anthropometric/social traits | ||||
| Beighton hypermobility | BHM | 23andMe | 18 | 64 |
| Breast size | CUP | 23andMe | 14 | 34 |
| Body mass index | BMI | 72 | 30 | 240 |
| Bone mineral density (femoral neck) | FNBMD | 17 | 19 | 33 |
| Bone mineral density (lumbar spine) | LSBMD | 17 | 21 | 32 |
| Chin dimples | DIMP | 23andMe | 57 | 58 / 13 |
| Educational attainment | EDU | 76 | 93 | 294 |
| Height | HEIGHT | 71 | 584 | 253 |
| Male pattern baldness | MPB | 23andMe | 49 | 9 / 8 |
| Nearsightedness | NST | 23andMe | 183 | 106 / 86 |
| Nose size | NOSE | 23andMe | 13 | 67 |
| Waist-hip ratio | WHR | 77 | 13 | 143 |
| Unibrow | UB | 23andMe | 61 | 69 |
|
Immune-related traits | ||||
| Any allergies | ALL | 23andMe | 43 | 67 / 114 |
| Asthma | ATH | 23andMe | 35 | 28 / 129 |
| Childhood ear infections | CEI | 23andMe | 15 | 47 / 75 |
| Crohn's disease | CD | 78 | 61 | 6 / 15 |
| Hypothyroidism | HTHY | 23andMe | 30 | 18 / 117 |
| Rheumatoid arthritis | RA | 79 | 74 | 14 / 44 |
| Tonsillectomy | TS | 23andMe | 48 | 60 / 113 |
| Ulcerative colitis | UC | 78 | 42 | 7 / 21 |
|
Metabolic phenotypes | ||||
| Age at menarche | AAM | 43 | 70 | 133 |
| Age at menarche (23andMe) | AAM (23) | 23andMe | 55 | 77 |
| Age at voice drop | AVD | 23andMe | 5 | 56 |
| Coronary artery disease | CAD | 45 | 11 | 22 / 65 |
| Type 2 diabetes | T2D | 80 | 11 | 12 / 57 |
| Fasting glucose | FG | 81 | 15 | 58 |
| Low-density lipoproteins | LDL | 82 | 41 | 85 |
| High-density lipoproteins | HDL | 82 | 46 | 89 |
| Triglycerides | TG | 82 | 31 | 86 |
| Total cholesterol | TC | 82 | 53 | 89 |
|
Hematopoeitic traits | ||||
| Hemoglobin | HB | 83 | 16 | 51 |
| Mean cell hemoglobin concentration | MCHC | 83 | 15 | 46 |
| Mean red cell volume | MCV | 83 | 42 | 48 |
| Packed red cell volume | PCV | 83 | 13 | 44 |
| Red blood cell count | RBC | 83 | 25 | 45 |
| Platelet count | PLT | 84 | 50 | 44 |
| Mean platelet volume | MPV | 84 | 29 | 17 |
Identification of genetic variants that influence pairs of traits
We first aimed to identify genetic variants that influence pairs of traits. To do this, we developed a statistical model (extending that used by Giambartolomei et al. 38) to estimate the probability that a given genomic region either 1) contains a genetic variant that influences the first trait, 2) contains a genetic variant that influences the second trait, 3) contains a genetic variant that influences both traits, or 4) contains both a genetic variant that influences the first trait and a separate genetic variant that influences the second trait (Figure 1). The input to the model is the set of summary statistics (effect size estimates and standard errors) for each SNP in the genome on each of the two phenotypes, and (if the two GWAS were performed on overlapping sets of individuals) the expected correlation in the summary statistics due to correlation between the phenotypes. We can then fit the following log-likelihood function:
where D is the data, M is the number of approximately independent blocks in the genome, Π0 is the prior probability that a region contains no genetic variants than influence either trait, Π1, Π2, Π3 and Π4 represent the prior probabilities of the four models described above, Θ is the set of all five Π parameters, and is the regional Bayes factor measuring the support for model j in genomic region i (see Supplementary Information for details). In the presence of missing data, we consider only the subset of SNPs with data in both studies; if the causal SNP is not present this acts to reduce power to detect a shared effect 38. In fitting this model, we estimate the prior parameters and the posterior probability of each model for each region of the genome (for numerical stability, in practice we penalize the estimates of the prior parameters, and so obtain maximum a posteriori estimates). We were mainly interested in the estimated prior probability that each genomic region contains a variant that influences both trait () and the corresponding posterior probabilities for each genomic region.
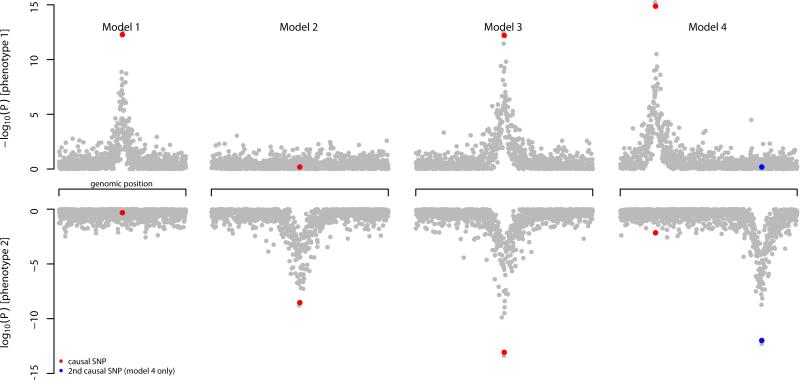
Figure 1. Schematic of the different models considered for a given genomic region and two GWAS.
We divide the genome into approximately independent blocks (see Methods), and estimate the proportion of blocks that fit into the shown patterns. The null model with no associations is not shown. Each point represents a single genetic variant.
Several caveats of this method are worth mentioning. First, note that the estimate is best thought of as the proportion of genomic regions that detectably influence both traits–if one study is small and underpowered, this estimate will necessary be zero. This contrasts with methods that aim to provide unbiased estimates of the “genetic correlation” between traits that do not depend on sample size 39-41. Second, in general it is not possible to distinguish a single causal variant that influences both traits (Model 3 in Figure 1) from two separate causal variants (Model 4 in Figure 1) in the presence of strong linkage disequilibrium between the causal variants. For any individual genomic region discussed below, the possibility of two highly correlated causal variants must be considered as an alternative possibility in the absence of functional follow-up. (Indeed, this latter possibility appears to be common in quantitative trait locus studies performed in model organisms 42). Finally, we evaluated the method in simulations (Supplementary Figures 1-5), and found that the model gives a small overestimate of proportion of shared effects (Supplementary Figure 3). This is because the amount of evidence against the null model of no associations is greater when a variant influences both phenotypes compared to when it only influence a single phenotype (Supplementary Figure 4).
Overlapping association signals identified in 43 GWAS
We applied the method to all pairs of the 43 GWAS listed in Table 1. For each pair of studies, we first estimated the expected correlation in the effect sizes from the summary statistics, and included this correction for overlapping individuals in the model. Note that this is conservative: in pairs of GWAS where we are sure there are no overlapping individuals (for example, age at menarche and age at voice drop) we see that the correlation in the summary statistics is non-zero, indicating that we are correcting out some truly shared genetic effects on the two traits (Supplementary Figure 6).
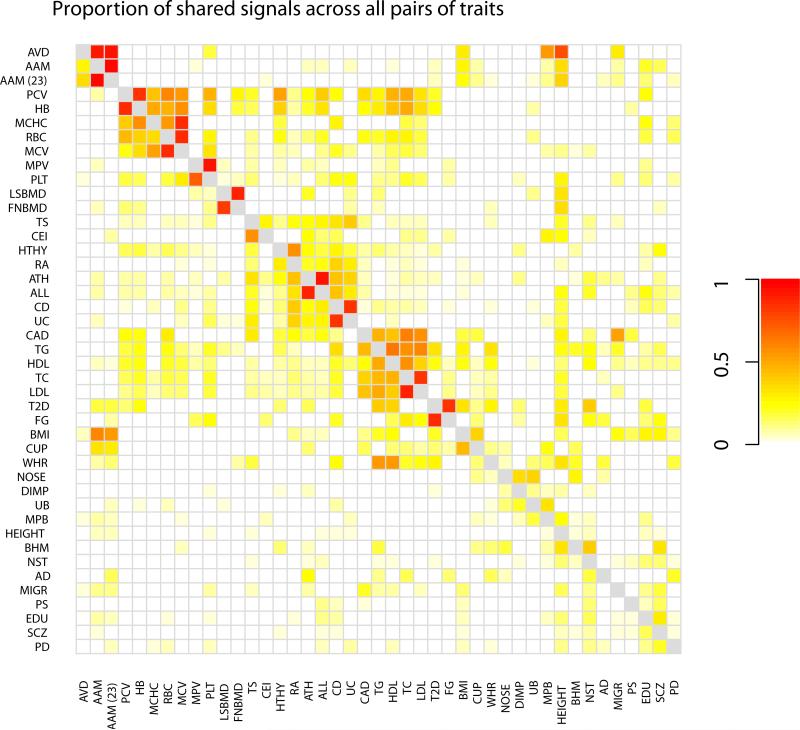
To gain an exploratory sense of the relationships between the phenotypes, we examined the patterns of overlap in associations among all 43 studies. Specifically, the model can be used to estimate, for each pair of traits [i,j], the proportion of detected variants that influence trait i that also detectably influence trait j. These estimates are shown in Figure 2, with phenotypes clustered according to their patterns of overlap. We see several clusters of related traits. For example, of the variants that detectably influence age at menarche (in the Perry et al. 43 study), the maximum a posteriori estimate is that 36% detectably influence height, 30% detectably influence age at voice drop, 28% influence BMI, 10% influence breast size, and 10% influence male pattern baldness. We interpret this as a set of phenotypes that share hormonal regulation. Additionally, there is a large cluster of phenotypes including coronary artery disease, type 2 diabetes, red blood cell traits, and lipid traits, which we interpret as a set of metabolic traits. Further, immune-related disease (allergies, asthma, hypothyroidism, Crohn's disease and rheumatoid arthritis) all cluster together, and also cluster with infectious disease traits (childhood ear infections and tonsillectomy). This biologically-revelant clustering validates the principle that GWAS variants can identify shared mechanisms underlying pairs of traits in a systematic way. As a control, we performed the same clustering of phenotypes by the estimated proportion of genomic regions where two causal sites fall nearby (Model 4 in Figure 1). In this case, there was no biologically-meaningful clustering (Supplementary Figure 7).
Figure 2. Heatmap showing patterns of overlap between traits.
Each square [i,j] shows the maximum a posteriori estimate of the proportion of genetic variants that influence trait i that also influence trait j, where i indexes rows and j indexes columns. Note that this is not symmetric. Darker colors represent larger proportions. Colors are shown for all pairs of traits that have at least one region in the set of 341 identified loci; all other pairs are set to white. Phenotypes were clustered by hierarchical clustering in R 74.
Individual loci that influence many traits
We next examined the individual loci identified by these pairwise GWAS. We identified 341 genomic regions where we infer the presence of a variant that influences a pair of traits, at a threshold of a posterior probability greater than 0.9 of model 3 (Supplementary Table 1). This number excludes “trivial” findings where a genetic variant influences two similar traits (two lipid traits, two red blood cell traits, two platelet traits, both measures of bone mineral density, both inflammatory bowel diseases, or type 2 diabetes and fasting glucose) and the MHC region. A previous “phenome-wide association study” identified 44 genetic variants associated with multiple phenotypes 34, so this represents an order-of-magnitude increase in the number of such loci.
Some genomic regions contain variants that influence a large number of the traits we considered. We ranked each genomic region according to how many phenotypes share genetic associations in the region (that is, if the pairwise scan for both height and CAD, and the pairwise scan for CAD and LDL, both indicated the same region, we counted this as three phenotypes sharing an association in the region). The top region in this ranking identified a non-synonymous polymorphism in SH2B3 (rs3184504) that is associated with a number of autoimmune diseases, lipid traits, heart disease, and red blood cell traits (Supplementary Figure 8; Supplementary Table 2). This variant has been identified in many GWAS, particularly for autoimmune disease 44.
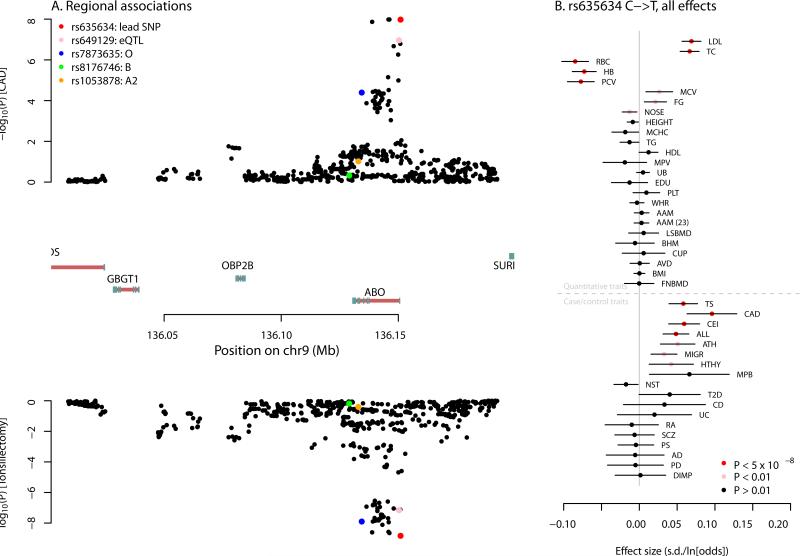
The next region in this ranking contains the gene coding for the ABO histo-blood groups in humans, and has a variant associated with 11 traits in these data (and many other additional traits not in these data, see also 20;45-47). In Figure 3A, we show the association statistics in this region for coronary artery disease and probability of having a tonsillectomy. At the lead SNP, the non-reference allele is associated with increased risk of CAD (Z = 5.7; P = 1.1 × 10−8) and increased risk of having a tonsillectomy (Z = 6.0; P =1.5 × 10−9). This variant is also strongly associated with other immune, red blood cell, and lipid traits in these data (Figure 3B). A tag for a microsatellite that influences the expression of ABO 48 is correlated to the lead SNP rs635634, as is a tag for the O blood group (Figure 3A). However, the lead SNP is an eQTL for both ABO and the nearby gene SLC2A6 in whole blood 46, so this allele may in fact have downstream effects via effects on the expression of two genes.
Figure 3. Multiple associations near the ABO gene. A. Association signals for coronary artery disease and tonsillectomy.
In the top panel, we show the P-values for association with coronary artery disease for variants in the window around the ABO gene. In the bottom panel are the P-values for association with tonsillectomy. In both panels, SNPs that tag functionally-important alleles at ABO are in color. In the middle are the gene models in the region–exons are denoted by blue boxes, and introns with red lines. Note that the ABO gene is transcribed on the negative strand. B. Association effect sizes for rs635634 on all tested traits. Shown are the effect size estimates for rs635634 for all traits. The lines represent 95% confidence intervals. Traits are grouped according to whether they are quantitative traits (in which case the x-axis is in units of standard deviations) or case/control traits (in which case the x-axis is in units of log-odds).
Among the top-ranked regions are several where the likely causal variant is known:
A non-synonymous variant in the zinc transporter SLC39A8 (rs13107325; Supplementary Figure 9) that is associated with schizophrenia (log-odds ratio of the non-reference allele = 0.15, P = 2 × 10−12), Parkinson's disease (log-odds ratio = −0.15, P = 1.6 × 10−7), and height s.d., P = 3.8 × 10−7), among others
A non-synonymous variant in the glucokinase regulator GCKR (rs1260326; Supplementary Figure 10) that is associated with fasting glucose ( s.d., P = 5 × 10−25) and height ( s.d., P = 2.6 × 10−11), among others.
A set of variants near the APOE gene (which we presume to be driven by the APOE4 allele; Supplementary Figure 11) that is associated with nearsightedness (rs6857 log-odds ratio = −0.04, P = 1.8 × 10−5), waist-hip ratio ( s.d., P = 8.3 × 10−5), and several lipid traits apart from the well-known association with Alzheimer's disease.
Regulatory variants in an intron of the FTO gene 49;50 that are associated with breast size in women (Supplementary Figure 12: rs1421085 s.d., P = 3.5 × 10−7) and age at voice drop in men ( s.d., P = 2.7 × 10−5), among others.
It has previously been observed that association signals for different phenotypes tend to cluster spatially in the genome 51; these results suggest that in some cases clustered associations are driven by single variants. We note anecdotally that the variants that influence a large number of phenotypes seem to often be non-synonymous, rather than regulatory, changes, which contrasts with the pattern seen in association studies overall (e.g. 37).
Identifying pairs of phenotypes with correlated effect sizes
In our scan for variants that influence pairs of phenotypes, we did not assume any relationship between the effect sizes of a variant on the two phenotypes. However, if two traits are influenced by shared underlying molecular mechanisms, we might expect the effects of a variant on the two phenotypes to be correlated. To test this, we returned to the set of variants identified by analysis of each phenotype individually (the numbers of these variants for each trait are in Table 1). For each set, we calculated the rank correlation between the effect sizes of the variants on the index trait (the one in which the variants were identified) and all of the other traits.
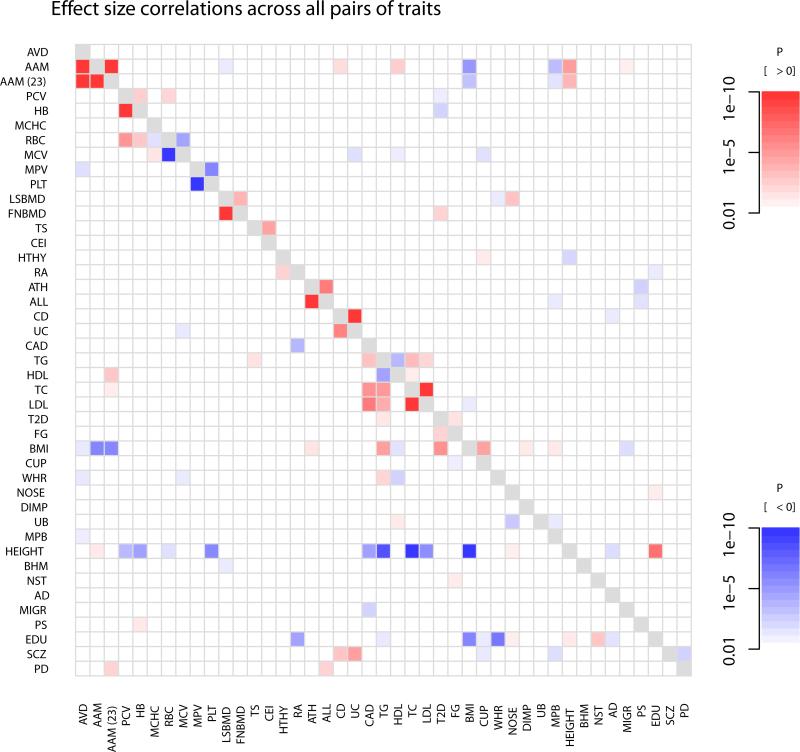
The results of this analysis are presented in Figure 4. Apart from closely related traits (e.g. the two measurements of bone density), we see a number of traits that are correlated at a genetic level. We focus on two of these. First, variants that delay age of menarche in women tend, on average, to decrease BMI (ρ = −0.53, P = 1.2 × 10−6), reduce risk of male pattern baldness (ρ = −0.45, P = 5.9 × 10−5), and increase height (ρ = 0.52, P = 2.2 × 10−6; Figure 4). These patterns hold both for the GWAS on age at menarche performed by Perry et al.43 and that performed by 23andMe (Figure 4). Most of these variants also delay age at voice drop in men (Figure 2), so we interpret these variants as ones that influence pubertal timing in general. The negative correlation between a variant's effect on age at menarche and BMI has previously been observed 39;43;52, as has the positive correlation between a variant's effect on age at menarche and height 39;43. The negative correlation between a variant's effect on age at menarche (or more likely, puberty in general) and male pattern baldness has not been previously noted, but is consistent with the known role for increased androgen signaling in causing hair loss 53-55.
Figure 4. Heatmap showing patterns of correlated effect sizes of variants across pairs of traits.
For each pair of traits [i,j], we extracted the set of variants that influence trait i and their effect sizes on both i and j. We then calculated Spearman's rank correlation between the effect sizes on i and the effect sizes on j, and tested whether this correlation was significantly different from zero. Shown in color are all pairs where this test had a P-value less than 0.01. Darker colors correspond to smaller P-values, and the color corresponds to the direction of the correlation (in red are positive correlations and in blue are negative correlations). The phenotypes are in the same order as in Figure 2. For a comparison to genome-wide genetic correlations, see Supplementary Figure 13.
Second, we find that genetic variants that increase risk of schizophrenia tend to increase risk of both Crohn's disease (ρ = 0.27, P = 2.2 × 10−4) and ulcerative colitis (ρ = 0.33, P = 6.6 × 10−6). These correlations (identified only at “significant” SNPs) are also present at the level of genome-wide genetic correlations between the diseases (39, Supplementary Figure 13). This observation is consistent with slightly higher rates of autoimmune diseases (including Crohn's and ulcerative colitis) in schizophrenia patients in Denmark 56-58, and with molecular evidence for a partial autoimmune etiology for schizophrenia (e.g. 59).
Inferring causal relationships between traits
Finally, we were interested in identifying pairs of traits may be related in a causal manner. Since we are using observational data (rather than, for example, a randomized controlled trial), we view strong statements about causality as impossible. Nonetheless, a realistic goal might be to identify aspects of the data that are more consistent with a causal model versus a non-causal model.
As a motivating example, we considered the correlation between levels of LDL cholesterol and risk coronary artery disease, now widely accepted as a causal relationship 60. We noticed that variants ascertained as having an effect on LDL cholesterol levels have correlated effects on risk of coronary artery disease (Figure 4, Figure 5C), while variants ascertained as having an effect on CAD risk do not in general have correlated effects on LDL levels (Figure 5D). This is consistent with the hypothesis that LDL cholesterol is one of many causal factors that influence CAD risk. An alternative interpretation is that LDL cholesterol is highly genetically correlated to an unobserved trait that causally influences risk of CAD.
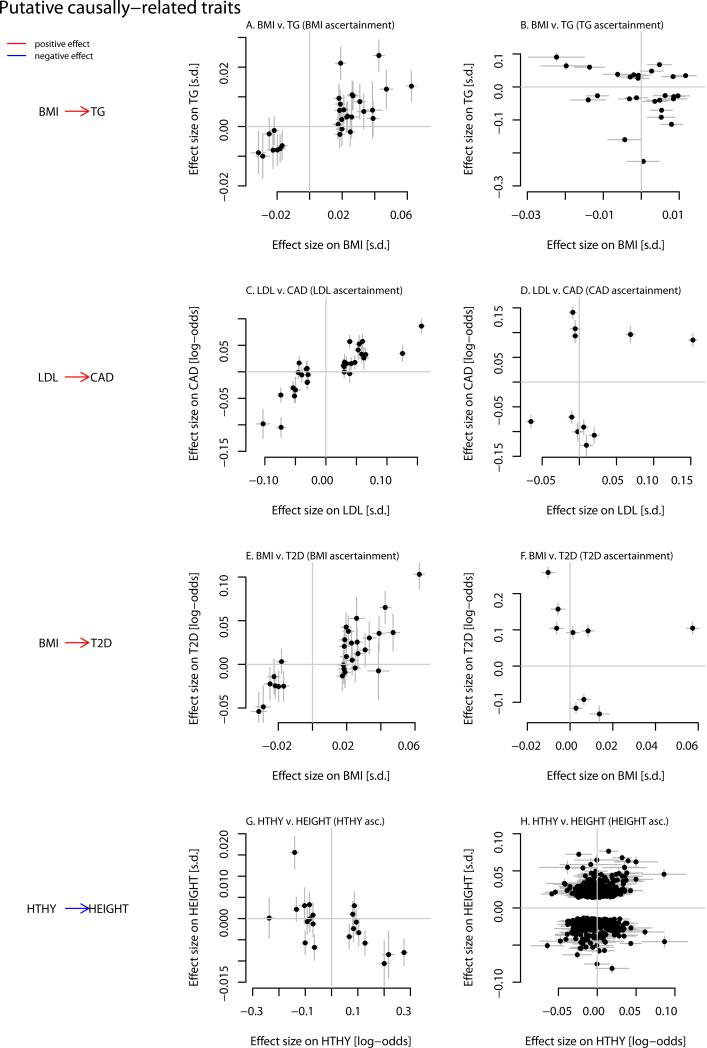
Figure 5. Putative causal relationships between pairs of traits.
For each pair of traits identified as candidates to be related in a causal manner (see Methods), we show the effect sizes of genetic variants on the two traits (at genetic variants successfully genotyped or imputed in both studies). Lines represent one standard error. A. and B. BMI and triglycerides. The effect sizes of genetic variants on BMI and triglyceride levels for variants identified in the GWAS for BMI (A.) or triglycerides (B.). C. and D. LDL and coronary artery disease. The effect sizes of genetic variants on LDL levels and coronary artery disease for variants identified in the GWAS for LDL (C.) or coronary artery disease (D.). E. and F. BMI and type 2 diabetes. The effect sizes of genetic variants on BMI and type 2 diabetes for variants identified in the GWAS for BMI (E.) or type 2 diabetes (F.). G. and H. Hypothyroidism and height. The effect sizes of genetic variants on hypothyroidism and height for variants identified in the GWAS for hypothyroidism (G.) or height (H.).
We developed a method to detect pairs of traits that show this asymmetry in the effect sizes of associated variants, which we interpret as more consistent with a causal relationship between the traits than a non-causal one (Methods). At a threshold of a relative likelihood of 100 in favor of a causal versus a non-causal model, we identified five pairs of putative causally-related traits. (At a less stringent threshold of a relative likelihood of 20 in favor of a causal model, we identified 11 additional pairs of traits (Supplementary Figure 14)) Simulations suggest this threshold corresponds approximately to a P-value around 0.001 (Supplementary Figure 15), and that the power of this test depends on the number of genetic variants used as input and the true underlying correlation in their effect sizes (Supplementary Figure 16). Four of these are shown in Figure 5. First, genetic variants that influence BMI have correlated effects on triglyceride levels, while the reverse is not true; this suggests increased BMI is a cause for increased triglyceride levels (Figure 5). Randomized controlled trials of weight loss are also consistent with this causal link 61;62, as are Mendelian randomization studies 63;64. Second, we confirm the evidence in favor of a causal role for increased LDL cholesterol in coronary artery disease (Figure 5), and in favor of a causal role for increased BMI in type 2 diabetes risk (Figure 5, Supplementary Figure 17). Finally, we suggest that increased risk of hypothyroidism causes decreased height (Figure 5). While it is known that severe hypothyroidism in childhood leads to decreased adult height (e.g. 65), these data indicate that hypothyroidism susceptibility may also influence height in the general population. A fifth potentially causal relationship (between risk of coronary artery disease and rheumatoid arthritis) could not be confirmed in a larger study and so is not displayed (see Supplementary Information, Supplementary Figure 18).
Discussion
We have performed a scan for genetic variants that influence multiple phenotypes, and have identified several hundred loci that influence multiple traits. This style of scan complements methods to quantify the “genetic correlation” between two traits 39;41;66;67 that are not generally concerned with identifying individual variants that influence both traits. We were interested in using the individual variants identified to identify biological relationships between traits, including potential relationships when one trait is causally upstream of the other. Other potential mechanisms that could lead to an association between a genetic variant and two phenotypes include trans-generational effects of a variant on a parental phenotype and a separate phenotype in the offspring (e.g. 68;69) or assortative mating that involves more than a single trait 70.
A number of limitations of this study are worth mentioning. First, all of the GWAS we have used are based on genotyping arrays and imputation, and so the loci identified are generally common (over 1% minor allele frequency). Inferences from common variants like these may not hold for rarer variants that may emerge from large sequencing studies. Second, we re-iterate that all of our inferences are based on sets of “detectable” loci; the GWAS we have used have highly variable sample sizes, and the traits have variable genetic architectures. As sample sizes for all traits reach the millions, inferences from “detectable” loci will converge to inferences from all loci. If traits truly follow an infinitesimal model (where every genetic variant influences every trait), we speculate that patterns of genetic overlap (like those in Figure 2) will become less interpretable, while patterns of genetic correlation (like those in Figure 4) may be more useful.
One clear observation from these data is that genetic variants that influence puberty (age at menarche and age at voice drop) often have correlated effects on BMI, height, and male pattern baldness (Figure 4). In our scan for causal relationships between traits, we found modest evidence of a causal role of age at menarche in influencing adult height, and for a causal role of BMI in the development of male pattern baldness (Supplementary Figure 12). The non-causal alternative (also consistent with the data) is that all of these traits are influenced by some of the same underlying biological pathways, and perhaps the most likely candidate is hormonal signaling. This highlights the importance of considering evidence from multiple traits when interpreting the molecular consequences of a variant and designing experimental studies. While variants that influence height overall are enriched near genes expressed in cartilage 71 and variants that influence BMI are enriched near genes expressed broadly in the central nervous system 72, it seems a subset of these variants also influence age at menarche and male pattern baldness. For these variants, it may be worth considering functional follow-up in gonadal tissues or specific brain regions known to be important in hormonal signaling.
It is also striking to note how many genetic variants influence multiple traits (Figure 2) but without a consistent correlation in the effect sizes (Figure 4). For example, many of the autoimmune and immune-related traits appear to share many genetic causes in common, but the effect sizes of the variants on the different traits appear to be largely uncorrelated (see also 10;39). Likewise, many variants appear to influence lipid traits, red blood cell traits and immune traits, but without consistent directions of effect. A trivial explanation of this observation is that we are underpowered to detect correlations in the effect sizes because we are using only a small set of the SNPs with the strongest associations. However, the genetic correlations between many of these traits (calculated using all SNPs) are not significantly different from zero (39, Supplementary Figure 13). Another possibility is that a given genetic variant often influences the function of multiple cell types through separate molecular pathways, or that the effects of a variant on two related phenotypes vary according to an individual's environmental exposures.
From the point of view of epidemiology, the ability to scan through many pairs of traits to find those that are potentially causally related seems appealing, and some previous analyses have had similar goals 73. Our approach makes the key assumption that, if two traits are related in a causal manner, then the “causal” trait is one of many factors that influence the “caused” trait. This induces an asymmetry in the effects of genetic variants on the two traits that can be detected (Figure 5). We also assume that we have identified a modest number of variants that influence both traits. This naturally means we are limited to considering heritable traits that have been studied with in cohorts with moderate sample sizes (on the order of tens to hundreds of thousands of individuals). It seems likely that the main limiting factor to scaling this approach (should it be generally useful) will be phenotyping rather than genotyping.
Methods
Methods are available in the Supplementary Materials.
Supplementary Material
Acknowledgements
This work was supported in part by the National Human Genome Research Institute of the National Institutes of Health (grant number R44HG006981 to 23andMe) and the National Institute of Mental Health (grant number R01MH106842 to JKP). We would like to thank the customers of 23andMe for making this work possible, the GWAS consortia that made summary statistics available to us, Luke Jostins for providing updated summary statistics from the Crohn's disease and ulcerative colitis GWAS, and Graham Coop and Matthew Stephens for helpful discussions. We thank David Golan, Jonathan Pritchard, and three anonymous reviewers for comments on a previous version of this manuscript. We thank David Cesarini and the Social Science Genetic Association Consortium for access to summary statistics from the association study of educational attainment.
Data on glycaemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. Data on coronary artery disease / myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG
We thank the International Genomics of Alzheimer's Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i-Select chips was funded by the French National Foundation on Alzheimer's disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Universit de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant 503480), Alzheimer's Research UK (Grant 503176), the Wellcome Trust (Grant 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer's Association grant ADGC-10-196728.
Footnotes
Author Contributions
JKP developed and applied the methods for pairwise analysis of association studies. TB contributed to the splitting of GWAS hits into independent blocks. JZL performed the LD score regression analyses. LS contributed to the analysis of the ABO region. JYT and DH performed and analysed the studies from 23andMe. All authors contributed to the writing of the manuscript.
URLs
gwas-pw code: https://github.com/joepickrell/gwas-pw
Approximately independent LD blocks: https://bitbucket.org/nygcresearch/ldetect-data
Competing Financial Interests
JYT and DH are employees of 23andMe, Inc.
References
- 1.Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–73. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29:66–73. doi: 10.1016/j.tig.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chillón M, et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. New England Journal of Medicine. 1995;332:1475–1480. doi: 10.1056/NEJM199506013322204. [DOI] [PubMed] [Google Scholar]
- 5.Müller C. Xanthomata, hypercholesterolemia, angina pectoris. Acta Medica Scandinavica. 1938;95:75–84. [Google Scholar]
- 6.Steinberg D. Atherogenesis in perspective: hypercholesterolemia and inflammation as partners in crime. Nat Med. 2002;8:1211–7. doi: 10.1038/nm1102-1211. [DOI] [PubMed] [Google Scholar]
- 7.Pearl J. Causality: models, reasoning and inference. Vol. 29. Cambridge Univ Press; 2000. [Google Scholar]
- 8.Steinberg D. The cholesterol controversy is over. why did it take so long? Circulation. 1989;80:1070–8. doi: 10.1161/01.cir.80.4.1070. [DOI] [PubMed] [Google Scholar]
- 9.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreassen OA, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreassen OA, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott KS, et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Ann Rheum Dis. 2013;72:935–41. doi: 10.1136/annrheumdis-2012-202081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivakumaran S, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89:607–18. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefansson H, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–6. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 16.Styrkarsdottir U, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–20. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 17.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moltke I, et al. A common greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–3. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 19.Pendergrass SA, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the population architecture using genomics and epidemiology (PAGE) network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, et al. Disease risk factors identified through shared genetic architecture and electronic medical records. Sci Transl Med. 2014;6:234ra57. doi: 10.1126/scitranslmed.3007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–8. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 22.Davey Smith G, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. International Journal of Epidemiology. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 23.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim ET, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panoutsopoulou K, et al. The effect of FTO variation on increased osteoarthritis risk is mediated through body mass index: a mendelian randomisation study. Ann Rheum Dis. 2013 doi: 10.1136/annrheumdis-2013-203772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes MV, et al. Causal effects of body mass index on cardiometabolic traits and events: a mendelian randomization analysis. Am J Hum Genet. 2014;94:198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Silva NMG, et al. Mendelian randomization studies do not support a role for raised circulating triglyceride levels influencing type 2 diabetes, glucose levels, or insulin resistance. Diabetes. 2011;60:1008–18. doi: 10.2337/db10-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granell R, et al. Effects of BMI, fat mass, and lean mass on asthma in childhood: a mendelian randomization study. PLoS Med. 2014;11:e1001669. doi: 10.1371/journal.pmed.1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichler I, et al. Serum iron levels and the risk of Parkinson disease: a mendelian randomization study. PLoS Med. 2013;10:e1001462. doi: 10.1371/journal.pmed.1001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 32.Fortune MD, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47:839–46. doi: 10.1038/ng.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis G, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasaniuc B, et al. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–14. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–73. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loh P-R, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–92. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–33. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry JRB, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–7. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4:6. doi: 10.1186/gm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wessel J, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franchini M, Lippi G. The intriguing relationship between the ABO blood group, cardiovascular disease, and cancer. BMC Med. 2015;13:7. doi: 10.1186/s12916-014-0250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kominato Y, Tsuchiya T, Hata N, Takizawa H, Yamamoto F. Transcription of human ABO histo-blood group genes is dependent upon binding of transcription factor CBF/NF-Y to minisatellite sequence. J Biol Chem. 1997;272:25890–8. doi: 10.1074/jbc.272.41.25890. [DOI] [PubMed] [Google Scholar]
- 49.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of gwas and age-associated diseases. Aging Cell. 2012;11:727–31. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elks CE, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li R, et al. Six novel susceptibility loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet. 2012;8:e1002746. doi: 10.1371/journal.pgen.1002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richards JB, et al. Male-pattern baldness susceptibility locus at 20p11. Nat Genet. 2008;40:1282–4. doi: 10.1038/ng.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamilton JB. Patterned loss of hair in man; types and incidence. Ann N Y Acad Sci. 1951;53:708–28. doi: 10.1111/j.1749-6632.1951.tb31971.x. [DOI] [PubMed] [Google Scholar]
- 56.Eaton WW, et al. Association of schizophrenia and autoimmune diseases: linkage of Danish national registers. Am J Psychiatry. 2006;163:521–8. doi: 10.1176/appi.ajp.163.3.521. [DOI] [PubMed] [Google Scholar]
- 57.Eaton W, et al. Coeliac disease and schizophrenia: population based case control study with linkage of Danish national registers. BMJ. 2004;328:438–9. doi: 10.1136/bmj.328.7437.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benros ME, et al. Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am J Psychiatry. 2011;168:1303–10. doi: 10.1176/appi.ajp.2011.11030516. [DOI] [PubMed] [Google Scholar]
- 59.Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scandinavian Simvastatin Survival Study Group Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S). Lancet. 1994;344:1383–9. [PubMed] [Google Scholar]
- 61.Look AHEAD Research Group et al. Reduction in weight and cardiovascular disease risk factors in individuals with type 2 diabetes: one-year results of the look ahead trial. Diabetes Care. 2007;30:1374–83. doi: 10.2337/dc07-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shai I, et al. Weight loss with a low-carbohydrate, Mediterranean, or low-fat diet. N Engl J Med. 2008;359:229–41. doi: 10.1056/NEJMoa0708681. [DOI] [PubMed] [Google Scholar]
- 63.Würtz P, et al. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med. 2014;11:e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freathy RM, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on bmi. Diabetes. 2008;57:1419–26. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivkees SA, Bode HH, Crawford JD. Long-term growth in juvenile acquired hypothyroidism: the failure to achieve normal adult stature. N Engl J Med. 1988;318:599–602. doi: 10.1056/NEJM198803103181003. [DOI] [PubMed] [Google Scholar]
- 66.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visscher PM, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10:e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueland PM, Hustad S, Schneede J, Refsum H, Vollset SE. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol Sci. 2001;22:195–201. doi: 10.1016/s0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- 69.Zhang G, et al. Assessing the causal relationship of maternal height on birth size and gestational age at birth: A mendelian randomization analysis. PLoS Med. 2015;12:e1001865. doi: 10.1371/journal.pmed.1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gianola D. Assortative mating and the genetic correlation. Theor Appl Genet. 1982;62:225–31. doi: 10.1007/BF00276244. [DOI] [PubMed] [Google Scholar]
- 71.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Evans DM, et al. Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9:e1003919. doi: 10.1371/journal.pgen.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R Core Team. R . A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org/ [Google Scholar]
- 75.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer's disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016 doi: 10.1038/nature17671. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature genetics. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gieger C, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.