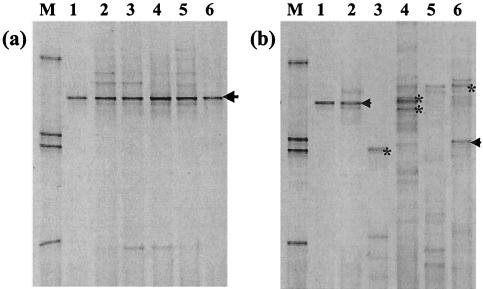
FIG. 2.
DGGE analysis of 16S rRNA gene sequences from various environmental samples amplified by nested PCR with primers 63F-665R and 357FGC-518R. (a) Deep marine sediment samples (Table 1). Lanes: M, DGGE marker (34); 1, clone Nank-B7, positive control; 2, ChCM core GeoB 7112-3; 3, ChCM core GeoB 7132-5; 4, ChCM core GeoB 7190-3; 5, Nankai Trough site 1173; 6, Peru Margin site 1229. (b) Other environmental samples (Table 1). Lanes: M, DGGE marker; 1, clone Nank-B7, positive control; 2, River Taff; 3, Chartley Moss; 4, Arabian Sea; 5, Cardiff University; 6, Arne Peninsula. Labeled DGGE bands represent bands that were excised and sequenced. JS1-like sequences are represented by arrows, and γ-proteobacteria are represented by asterisks.