FIG.1.
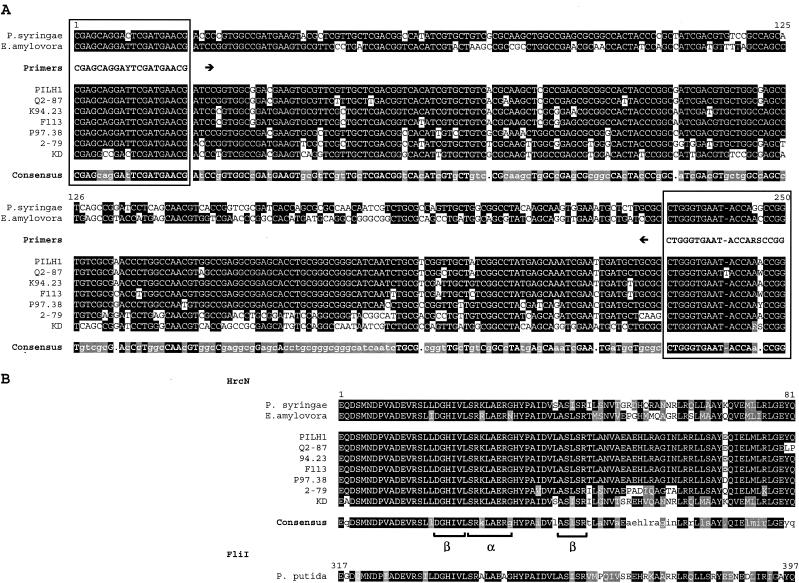
Alignment of partial hrcN nucleotide (A) and deduced HrcN amino acid (B) sequences of P. syringae pv. tomato DC3000, E. amylovora CNPB136, and selected biocontrol pseudomonads, including PILH1 (hrcN group 1), Q2-87 (hrcN group 2), K94.23 (hrcN group 3), F113 (hrcN group 4), P97.38 (hrcN group 6), 2-79 (hrcN group 8), and KD (hrcN group 10). The sites annealing to the PCR primers hrcN-4r (reverse) and hrcN-5rR (forward) are enclosed in boxes. The hrcN consensus sequence is indicated by uppercase letters (>90% identity), lowercase letters (between 50 and 90% identity), or a dot (<50% identity). The HrcN consensus sequence is indicated by a black background (100% identity) or a grey background (<100% identity but 100% similarity). Similarity was calculated by using a BLOSUM62 matrix (23) implemented in LALIGN (26). For each strain, amino acids are indicated by a black background if they are identical to the amino acids in the consensus sequence or by a grey background in case of homologous amino acids. The deduced FliI sequence of P. putida KT2440 is shown for reference.
