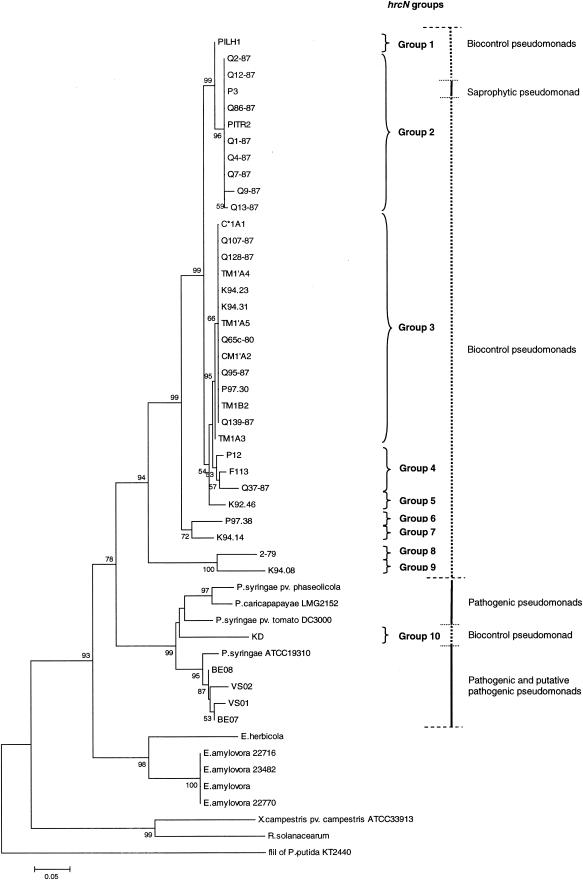
FIG.2.
Phylogenetic relationships based on partial hrcN sequences of biocontrol fluorescent pseudomonads and phytopathogenic bacteria belonging to the genera Pseudomonas, Erwinia, Xanthomonas, and Ralstonia. The distance tree was generated by the NJ method with the JC formula by using the flagellar ATPase gene fliI of P. putida KT2440 (accession number AE016790) as the outgroup. Nodal support was assessed by using 1,000 bootstrap replicates. Only bootstrap values greater than 50% are shown. Scale bar = 0.05 substitution per site. hrcN groups arbitrarily defined for biocontrol pseudomonads based on the topology of the hrcN tree are indicated on the right. The phytopathogenic bacteria included P. syringae pv. phaseolicola (accession number AJ430232), P. syringae pv. tomato DC3000 (AE016860), Erwinia herbicola (X99768), E. amylovora (L25828), X. campestris pv. campestris ATCC 33913 (AE012222), and Ralstonia solanacearum (AJ245811). Sequences were obtained in this study for Pseudomonas strains KD (AY456994), VS01 (AY456998), VS02 (AY456999), BE07 (AY457000), BE08 (AY457001), K94.23 (AY457002), C*1A1 (AY457003), TM1′A4 (AY457004), PITR2 (AY457005), K94.31 (AY457006), Q65c-80 (AY457007), CM1′A2 (AY457008), TM1′A5 (AY457009), Q1-87 (AY457010), TM1B2 (AY457011), Q7-87 (AY457012), Q86-87 (AY457013), Q9-87 (AY457014), Q12-87 (AY457015), Q128-87 (AY457016), P97.30 (AY457017), Q139-87 (AY457018), TM1A3 (AY457019), P97.38 (AY457020), F113 (AY457021), Q95-87 (AY457022), Q37-87 (AY457023), and 2-79 (AY457024), Pseudomonas caricapapayae LGM2152 (AY457025), Pseudomonas strains Q4-87 (AY457026), Q2-87 (AY457027), P12 (AY457028), P3 (AY457029), K92.46 (AY457030), K94.14 (AY457031), Q107-87 (AY457032), Q13-87 (AY457033), K94.08 (AY457034), and PILH1 (AY457035), P. syringae ATCC 19310 (AY457036), and E. amylovora 22716 (AY456995), 22770 (AY456996), and 23482 (AY456997). The G+C contents of the hrcN fragment studied were 58.3% ± 2.5% for the Erwinia cluster (n = 5), 60.4% ± 2.0% for the cluster comprised of established and putative pathogenic pseudomonads and strain KD (n = 9) (the G+C content of KD was 60.6%), and 61.7% ± 1.0% for the cluster containing all biocontrol pseudomonads except strain KD and the saprophytic pseudomonad P3 (n = 33). The values for the latter two clusters were significantly different (P < 0.05, as determined by the Mann-Whitney test).