Abstract
Two different strains, Aspergillus awamori TGDTh-4 and A. awamori TGP-3 overexpressing a synthetic gene encoding the plant sweet protein thaumatin, showed an unfolded protein response. To facilitate protein secretion, the chaperone BiPA gene was expressed in A. awamori under control of the strong constitutive promoter of the gpdA gene. A good correlation was observed between the level of the bipA transcript in different strains and the amount of thaumatin secreted. Thaumatin secretion was increased 2- to 2.5-fold in transformants overexpressing the bipA gene compared with the parental strain. Secretion of the homologous proteins α-amylase and glucoamylase was not affected by the bipA gene overexpression. The requirement for BiPA for secretion of thaumatin was confirmed by attenuation of the endogenous bipA gene expression with an antisense RNA cassette. The decrease in bipA expression reduced the amount of secreted thaumatin up to 80% without affecting the secretion of the homologous α-amylase and glucoamylase proteins. The BiPA protein is, therefore, very important for secretion of some heterologous proteins, such as thaumatin in A. awamori.
One of the main bottlenecks in the production of heterologous proteins in filamentous fungi (11) is the secretion of the proteins. Protein traffic through the secretory pathway depends upon the levels of chaperones and foldases that play an important role in protein folding during secretion (1, 5). It has been observed previously that bottlenecks in the secretory pathway of Aspergillus awamori result in partial intracellular retention of heterologous proteins (4, 18). However, the molecular mechanism underlying these bottlenecks remains largely unknown.
The BiPA protein is a molecular chaperone that contains an HDEL retention signal and is present in the endoplasmic reticulum (2, 24). BiPA plays an important role in the protein traffic process by binding to the newly synthesized polypeptides and promoting their proper folding. It also binds to aberrant proteins, preventing them from leaving the endoplasmic reticulum to continue through the secretory pathway (26). Indeed, the BiPA protein appears to have a quality control function, discriminating between properly folded proteins to be exported (secretion competent) and inadequately folded proteins (12). The bipA gene of A. awamori was cloned (14) in order to study its role in protein secretion.
Thaumatin is an extremely sweet protein derived from the tropical plant Thaumatococcus daniellii (40). Its large-scale production is of great interest because of its use as a human food sweetener and as an animal feed supplement (8). The 22-kDa thaumatin protein contains eight disulfide bonds, and proper folding is required for the sweet taste. A synthetic thaumatin gene (tha) has been expressed previously in A. awamori (18, 19), but part of the thaumatin is retained intracellularly, indicating that it does not proceed adequately through the protein secretory pathway. Overexpression of heterologous proteins in filamentous fungi frequently results in an unfolded protein response (UPR) that leads to increased synthesis of chaperones (23, 36).
It has been observed previously that a defined level of the foldase protein disulfide isomerase (PDI) is required for optimal secretion of thaumatin by A. awamori (21). However, an excess of PDI in the cell reduced thaumatin production. It is likely that foldases and other chaperones have distinct roles in folding and secretion of different proteins.
In order to understand the role of BiPA in homologous and heterologous protein secretion, it was of interest to study the effect of different BiPA levels in A. awamori. These different levels could be obtained by overexpression and attenuation of the bipA gene.
In this paper we show that overexpression of the bipA gene in A. awamori causes an increase in the secreted levels of the heterologous protein thaumatin and that there is an optimal level of bipA expression, above which the secreted levels of thaumatin decrease. On the other hand, attenuation of the bipA gene causes a decrease in the secreted levels. Homologous proteins, such as α-amylase and glucoamylase, are not affected in any case.
MATERIALS AND METHODS
Strains.
A. awamori TGDTh-4, containing the tha gene under control of the nitrogen-regulated gdhA (glutamate dehydrogenase) promoter (18), was used as a host strain for bipA gene overexpression studies. A. awamori strain TGP-3, which produces thaumatin under control of the constitutive gpdA (glyceraldehyde-3-phosphate dehydrogenase) promoter, was used as a host strain for bipA gene downregulation studies. Both strains were derived from the mutant A. awamori lpr66, which is defective in aspergillopepsin A (19). Escherichia coli DH5α served as a host for plasmid amplification and purification.
Media and growth conditions.
All A. awamori strains were grown on solid Power sporulation medium (9) at 30°C for 3 days. For isolation of total DNA, A. awamori spores were inoculated into CM medium (5 g of malt extract per liter, 5 g of yeast extract per liter, 5 g of glucose per liter), and the cultures were grown at 30°C for 48 h in a rotary shaker at 300 rpm.
For thaumatin, α-amylase, or glucoamylase production studies, A. awamori seed cultures in CM medium were used to inoculate (18%, vol/vol) 100 ml of MDFA medium (18, 19) in 500-ml flasks and grown at 30°C for 60 h in a rotary shaker at 300 rpm. Culture samples for transcriptional analysis were collected after 48 h of incubation in MDFA medium.
Plasmids and plasmid construction.
Plasmid pGpd-BiP-H (containing the expression cassette shown in Fig. 1A) was used for overexpression of the bipA gene. This plasmid contains the A. awamori bipA gene under control of the Aspergillus nidulans gpdA promoter (PgpdA) and the pepB gene transcriptional terminator of A. awamori (TpepB). The hygromycin resistance gene (hph) from E. coli (under control of the A. nidulans gpdA promoter [PgpdA] and the trpC gene transcriptional terminator [TtrpC] of the same organism) was used as a selectable marker. A 2.2-kb fragment of genomic DNA corresponding to the open reading frame of the bipA gene was amplified from plasmid pB4S5-SK+ (14) by PCR by using the following oligonucleotides as primers: ObipA (5′-TTTTCCATGGCGCGCATCTCGCATCAG-3′), which corresponded to positions 1 to 21 of the bipA gene and introduced an NcoI site into its 5′ end; and ObipB (5′-TTACAGTTCATCATGCCCGCT-3′), which corresponded to positions 2168 to 2189 of the gene.
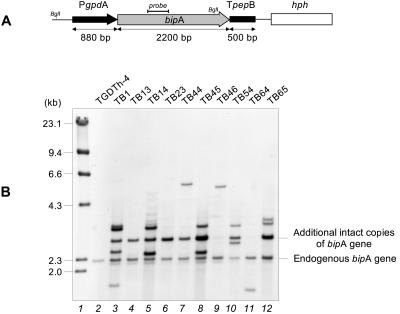
FIG. 1.
Overexpression cassette of the bipA gene of A. awamori and Southern blot analysis of its expression. (A) The bipA gene (gray arrow) is under control of the A. nidulans gpd promoter (PgpdA) (black arrow) and the A. awamori aspergillopepsin B transcriptional terminator (TpepB) (black box). The hph gene (open box) from E. coli was used as a selectable marker. (B) Total DNA was digested with BglI and hybridized with a 400-bp NcoI fragment close to the 5′ end of the bipA gene as a probe (solid line in panel A). Lane 1, size markers (λ HindIII fragments); lane 2, control strain TGDTh-4; lanes 3 to 12, A. awamori transformants expressing the bipA gene.
The PCR-amplified fragment was digested with the restriction enzymes StuI and NcoI and was cloned in plasmid pIBFM4 (20). The plasmid obtained, designated pGpd-BiP, was digested with the ApaI and XbaI restriction enzymes to obtain the bipA overexpression cassette, which was subcloned in vector pAN7-1Z (20) carrying the hygromycin resistance gene to obtain plasmid pGpd-BiP-H.
For the bipA gene attenuation studies, plasmid pGdhBiPAS3 (containing the antisense expression cassette shown below [see Fig. 5A]) was constructed. This plasmid contained a fragment of the bipA gene of A. awamori in the antisense position under control of the A. awamori glutamate dehydrogenase promoter (PgdhA) and the transcriptional terminator region of the cyc1 gene of Saccharomyces cerevisiae. The hygromycin resistance cassette was used as a selectable marker.
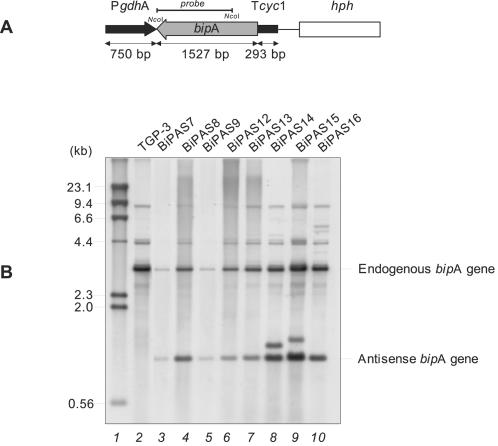
FIG. 5.
Downregulation of the bipA gene of A. awamori. (A) A fragment of the bipA gene (gray arrow), in the antisense position, is under control of the A. awamori gdh promoter (PgdhA) (black arrow) and the S. cerevisiae transcriptional terminator of the cyc1 gene (Tcyc1) (black box). The hph gene (open box) from E. coli was used as a selectable marker. (B) Southern blot analysis of transformants containing the bipA antisense cassette. Total DNA was digested with NcoI and hybridized with a 1,100-bp NcoI fragment of the bipA gene as a probe (solid line in panel A). Lane 1, size markers (λ HindIII fragments) (sizes are indicated on the left); lane 2, A. awamori control strain TGP-3, containing a tha expression cassette; lanes 3 to 10, A. awamori bipA antisense transformants.
The bipA gene used in the antisense construction included a 1,526-bp fragment of cDNA corresponding to the bipA gene except for the first 490 bp of the 5′ end. This fragment was amplified by PCR by using plasmid pMar (which contained the bipA gene in a pBluescript vector [Lombraña and Martín, unpublished data]) as the template and oligonucleotides ObipAS1 (5′GTTTCCGCCCGGGTTCTTGGT-3′), corresponding to positions 605 to 626 of the bipA gene, and ObipAS2 (5′-CAGTTCACCATGGCCGCTGG-3′), corresponding to positions 2166 to 2186 of the gene, as primers. The latter primer introduced an NcoI site into the 3′ end of the amplified fragment.
The cloned PCR product was used to replace an NcoI-StuI fragment containing the ble gene of plasmid pIBRC43 (3) to create pGdhBiPAS2, which carried the bipA gene in the antisense position. The antisense cassette was subcloned from pGdhBiPAS2 into pAN71 (27) (digested with Eco147I-XhoI) to obtain pGdhBiPAS3.
Transformation of A. awamori.
Protoplasts of A. awamori strains were transformed by the polyethylene glycol method (41). Hygromycin-resistant transformants were selected in tryptic soy agar containing 1 M sorbitol as an osmotic stabilizer supplemented with 200 μg of hygromycin per ml.
DNA purification and Southern hybridization.
Extraction and purification of total A. awamori DNA were performed as described previously (18, 19).
Southern analysis was performed by standard techniques (33). To analyze transformants overexpressing the bipA gene, a 400-bp NcoI fragment close to the amino-terminal end of bipA was used as a probe, and the total DNA of the different transformant and control strains were digested with BglI (Fig. 1A). Transformants containing the antisense bipA gene were probed with a 1.1-kb NcoI fragment of the bipA antisense construct; total DNA was digested with NcoI (see Fig. 5A). All probes were labeled with digoxigenin by the random priming method by following the instructions of the manufacturer (Roche Diagnostics GmbH, Mannheim, Germany).
The nylon filter was hybridized overnight at 42°C in a buffer containing 40% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sarcosine, 0.02% sodium dodecyl sulfate (SDS), and 2% blocking reagent. The filter was washed once with 2× SSC-0.1% SDS at 42°C for 15 min, once with 0.1× SSC-0.1% SDS at 42°C for 15 min, and once more with 0.1× SSC-0.1% SDS at 65°C for 15 min. Digoxigenin detection was performed as described by the manufacturer (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom).
RNA isolation and purification.
For RNA isolation, mycelia of A. awamori strains, frozen in liquid nitrogen, were ground to a fine powder in a mortar. The frozen powdered mycelium (50 mg for each sample) was mixed with 1.3 ml of EFA buffer (1 volume of extraction buffer, 1 volume of phenol, and 7 μl of β-mercaptoethanol per ml) and mixed for 1 min with a vortex mixer. The extraction buffer contained 4 M guanidinium isothiocyanate and 0.5% (wt/vol) N-laurylsarcosine in 25 mM sodium citrate buffer (pH 7.0). The mixture was centrifuged at 16,000 × g for 5 min at 4°C. The supernatant was collected, and 130 μl of chloroform-isoamyl alcohol (24:1) was added to each tube. After shaking, the tubes were centrifuged again at 16,000 × g at 4°C for 5 min. The upper layer was collected and extracted with 1 volume of phenol-chloroform and then with 1 volume of chloroform.
The RNA was precipitated with 0.75 volume of 8 M LiCl for 30 min at 4°C, and the precipitate was collected by centrifugation at 16,000 × g and 4°C for 15 min. After two washes with 70% (vol/vol) ethanol, the RNA was redissolved in water treated with diethyl pyrocarbonate. The mRNA was purified from the total RNA preparation by using an Oligotex mRNA kit (QIAGEN, Crawley, United Kingdom).
Northern analysis.
Purified mRNA (5 μg) was electrophoresed on a 1.2% (wt/vol) agarose formaldehyde gel at 87 V for 4 h. The gel was blotted onto a nylon filter (Zeta probe GT membrane; Bio-Rad, Richmond, Calif.) by standard methods, and the RNA was fixed by UV irradiation with a UV-Stratalinker 2400 lamp (Stratagene, La Jolla, Calif.). The filter was prehybridized for 4 h at 50°C in high-SDS buffer (7% SDS, 50% formamide, 5× SSC, 2% blocking reagent, 50 mM sodium phosphate [pH 7.0], 0.1% N-laurylsarcosine) and then hybridized at 50°C for 14 h in the same buffer containing the labeled probe. A 1.9-kb BamHI fragment corresponding to the complete cDNA of the bipA gene was used as the probe. A 0.83-kb NcoI-KpnI fragment of the A. nidulans β-actin gene was used as the probe in control hybridizations.
After hybridization, the filters were washed twice in 2× SSC-0.1% SDS for 30 min, once in 1× SSC-0.1% SDS for 30 min, and once in 0.2× SSC-0.1% SDS at 55°C for 45 min.
RT-PCR amplification.
To detect antisense transcripts of the bipA gene, reverse transcription (RT)-PCR was performed with the Superscript One Step system (Invitrogen, Carlsbad, Calif.). Purified mRNA of transformants and control strain TGP-3 grown in MDFA medium for 48 h were used as templates. The primers used are shown in Table 1. The program used for cDNA amplification included a reverse transcription step at 45°C for 40 min and 94°C for 2 min, followed by a PCR consisting of 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1.4 min and a final extension step at 72°C for 10 min.
TABLE 1.
Primers used for amplification of sense and antisense bipA RNA
| Fragment amplified | Oligonucleotide | Sequence | Position in gene |
|---|---|---|---|
| bipA transcript (986 bp) | ORNABiPAS1 | 5′-TTCCGCCATGGTTCTTG-3′ | 608-621 |
| ORNABiPAS2 | 5′-TTGGTCAGGGAACGCTCA-3′ | 1631-1648 | |
| Antisense bipA RNA | ORNABiPAS1 | 5′-TTCCGCCATGGTTCTTG-3′ | 608-621 |
| Opgdha | 5′-TTCCTGTTTTCCTAGATACCCCA-3′ |
The Opgdh oligonucleotide corresponds to the nucleotide sequence between the CTATA and CAAT boxes of the gdhA promoter.
Extraction of intracellular proteins.
To isolate the intracellular proteins, washed mycelia of A. awamori strains were frozen in liquid nitrogen and ground in a mortar. The frozen powdered mycelium (100 mg for each sample) was mixed with extraction buffer (625 mM Tris buffer [pH 6.5] containing 1 mM dithiothreitol, 1 μM phenylmethylsulfonyl fluoride, and 8 μl protease inhibitor cocktail [Sigma, St. Louis, Mo.] per ml).
Proteases in the protein mixtures were inactivated at 100°C for 15 min (to avoid proteolytic processing of BiPA) and then were centrifuged at 16,000 × g and 4°C for 20 min. The supernatants containing the intracellular proteins were collected and kept at −20°C until they were used.
Immunoblot analysis of A. awamori culture supernatants and intracellular proteins.
After electrophoresis of proteins on SDS-polyacrylamide gels, the proteins were transferred to a polyvinylidene difluoride membrane (Inmobilon-P; Millipore Corp. Bedford, Mass.) by using a Minitransblot electroblotting system (Bio-Rad). The membranes were treated with (i) anti-thaumatin polyclonal antibodies (serum dilution, 1:15,000), (ii) anti-glucoamylase polyclonal antibodies (serum dilution, 1:30,000), (iii) anti-PDI polyclonal antibodies (serum dilution, 1:10,000), or (iv) anti-BiP polyclonal antibodies (serum dilution, 1:100,000) in 50 mM Tris-HCl (pH 8.0)-150 mM sodium chloride containing Tween 20 at a concentration of 0.2% (vol/vol) for 1 h and then for 30 min with anti-rabbit alkaline phosphatase conjugate (1:10,000; Sigma) in the same buffer. The membranes were treated with a BCIP (5-bromo-4-chloro-3-indolylphosphate)—nitroblue tetrazolium substrate for alkaline phosphatase until the color was developed.
Anti-PDI and anti-glucoamylase antibodies were provided by D. Archer and D. Jeenes. Anti-BiP and anti-thaumatin antibodies were developed in our laboratory.
Quantification of α-amylase activity and thaumatin in A. awamori culture supernatants.
α-Amylase activity was measured by the method of García-González et al. (10). One unit of activity was defined as the amount of enzyme that released 1 nmol of maltose per s at 37°C and pH 7.2, and the specific activity was expressed in units per milligram of protein. Thaumatin was quantified by an enzyme-linked immunosorbent assay (ELISA) method as described previously (18).
Densitometric analysis.
bipA expression levels and thaumatin secretion levels were measured by densitometry of the secreted thaumatin and bipA gene expression signals by using the software Science Lab 2003 (Fuji Photo Film Co., Ltd., Tokyo, Japan).
RESULTS
Overexpression of the bipA gene in A. awamori.
To test the effect of the BiPA protein on secretion of heterologous and homologous proteins in A. awamori, an expression cassette containing the bipA gene under control of the constitutive promoter PgpdA of A. nidulans (Fig. 1A) was introduced into the thaumatin-producing strain A. awamori TGDTh-4 (18).
Ten transformants were randomly selected for Southern analysis (Fig. 1B). This analysis revealed the presence of a 2.3-kb fragment corresponding to the endogenous gene which was observed in both transformant and control strains. Another band at 3.0 kb, corresponding to the intact additional copies of the gene, was observed in most of the transformants.
The intensity of the 3.0-kb hybridization fragment in different transformants, compared with the intensity of the endogenous bipA gene, indicated that there was a different copy number of the gene in each transformant. Other bands corresponding to noncharacterized integration events were also observed.
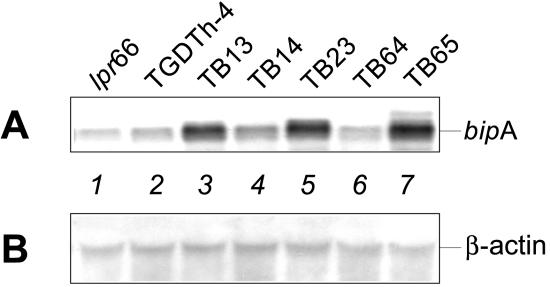
The transformants that exhibited different Southern hybridization patterns were selected for transcriptional analysis of the bipA gene. Northern blot analysis (Fig. 2) showed that there was an increase in the bipA mRNA level in some of the transformants (particularly strains TB13, TB23, and TB65) compared with the bipA transcript levels observed in the TGDTh-4 and lpr66 control strains; lpr66 did not contain the tha gene (it was a nonproducing strain) (Fig. 3).
FIG. 2.
(A) Northern blot analysis of transformants overexpressing the bipA gene. Hybridization was performed with a 1,900-bp BamHI probe of the bipA gene. Lane 1, control strain lpr66 (untransformed); lane 2, control strain TGDTh-4, containing the tha expression cassette; lanes 3 to 7, A. awamori transformants overexpressing the bipA gene. (B) Control Northern blot analysis of the same RNA samples performed with a 0.83-kb NcoI-KpnI fragment of the constitutive A. nidulans β-actin gene as the probe.
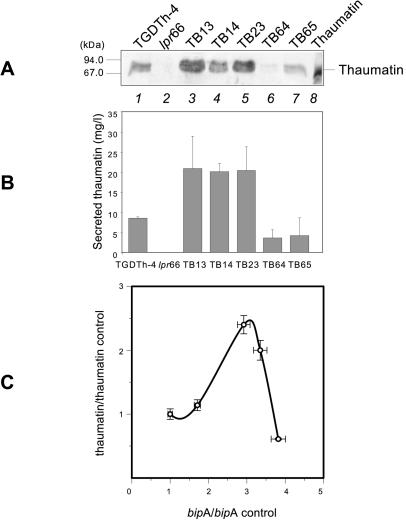
FIG. 3.
Secreted thaumatin in control strains and A. awamori transformants overexpressing the bipA gene. (A) Immunoblots with anti-thaumatin antibodies (1:15,000). Lane 1, control strain TGDTh-4, containing the tha expression cassette; lane 2, control strain lpr66 (untransformed); lanes 3 to 7, A. awamori TB transformants containing the bipA overexpression cassette; lane 8, purified 22-kDa thaumatin from T. daniellii (50 ng). The molecular masses of size markers are indicated on the left. (B) Levels of extracellular thaumatin measured by ELISA in the A. awamori control strain TGDTh-4 and in transformants overexpressing the bipA gene. The error bars indicate standard deviations for three measurements. (C) Correlation between thaumatin secretion and bipA expression levels in the control strain and different A. awamori transformants overexpressing the bipA gene. The data are expressed in units relative to the control strain levels. From left to right the data points correspond to the following strains: TGDTh-4, TB14, TB13, TB23, and TB65. Note that the thaumatin secretion levels depend strongly on the bipA expression levels.
Transformant TB64, which did not contain any additional intact copies of the bipA gene, did not exhibit an increase in the bipA transcript level compared to parental strain TGDTh-4.
An interesting observation was made when we compared the levels of bipA mRNA in the TGDTh-4 and lpr66 control strains. As shown in Fig. 2A, expression of the thaumatin-encoding gene in strain TGDTh-4 resulted in levels of bipA mRNA that were 30% higher than those in parental strain lpr66, presumably due to the UPR that is known to enhance the chaperone levels in response to accumulation of unfolded exogenous proteins.
Effect of bipA gene overexpression on secretion of thaumatin.
Transformants TB13, TB14, and TB23 (Fig. 3A) showed higher levels of secreted thaumatin than the control TGDTh-4 strain; nevertheless, transformants TB64 and TB65 showed levels that were even lower than the level in this control strain, and strain lpr66 showed no thaumatin signal, as expected. There was a nonlinear correlation with bipA gene expression intensity since these transformants showed high levels of the bipA transcript (Fig. 2). Transformant TB64, which did not produce the band corresponding to the intact additional copies of the bipA gene in the Southern analysis, showed a very faint thaumatin signal in the Western analysis. This fact could be explained by the partial or total integration of the bipA expression vector at the site of the thaumatin expression cassette, which could have caused a rearrangement that decreased the thaumatin production in this transformant strain. Alternatively, an alteration or a disruption of the thaumatin expression cassette could have been caused by integration of the bipA expression vector, which also would have caused a reduction in thaumatin secretion.
Results similar to the results obtained in the Western blot analysis were observed when thaumatin was quantified by an ELISA method (Fig. 3B). Transformants TB13, TB14, and TB23 showed a maximal twofold increase in the extracellular thaumatin level compared to parental strain TGDTh-4.
The quantitative correlation between bipA gene expression and thaumatin secretion is shown in Fig. 3C. As described previously for other foldases (21), a defined level of bipA expression is required for optimal secretion of thaumatin, and lower and higher levels of bipA expression cause decreases in secreted thaumatin levels. Transformant TB64 did not follow the same trend as the rest of transformants, presumably because of its different integration pattern, which could have caused rearrangements or even disruption at the site of the thaumatin expression cassette, as explained above.
Intracellular BiPA and PDI protein levels.
The levels of intracellular BiPA protein in transformants and control strains were analyzed with anti-BiP antibodies. Surprisingly, the BiPA protein was extremely unstable following cell disruption, and several immunoreactive bands were observed in the Western blot analysis (data not shown), suggesting that there was rapid proteolytic degradation of BiPA. Transformants TB13, TB14, and TB23 showed higher intensity of the immunoreactive bands, but, due to the degradation, it was difficult to quantify the real BiPA levels in the different strains.
In order to study whether overexpression of the bipA gene had any effect on the PDI chaperone, intracellular protein samples were analyzed by Western blotting with anti-PDI antibodies (data not shown). There were no differences between transformant strains and the control TGDTh-4 strain. Nevertheless, the PDI levels in strain lpr66 (which does not produce thaumatin) were much lower than those in the TGDTh-4 strain. These results indicate again that there was an UPR effect on PDI levels when the heterologous protein thaumatin was overexpressed in the TGDTh-4 strain.
Effect of bipA gene overexpression on secretion of homologous proteins.
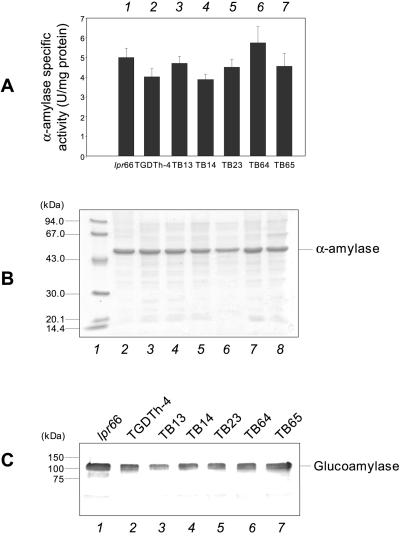
To test the effect of bipA gene overexpression on secretion of homologous proteins, the α-amylase activity was measured in the culture supernatants of the different A. awamori strains, and this protein was also identified by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 4). No apparent differences were found in the α-amylase activity values (Fig. 4A) between transformants overexpressing the bipA gene and control strains or in the α-amylase protein band intensities in SDS-PAGE gels (Fig. 4B). The α-amylase band was the major protein band in A. awamori culture supernatants and was identified in the gels by using pure α-amylase. Similar results were obtained when the secreted glucoamylase, another homologous protein, was visualized by Western blot analysis (Fig. 4C) in control and transformant strains.
FIG. 4.
Levels of homologous extracellular enzymes in A. awamori transformants. (A) Extracellular α-amylase activities of control strains lpr66 and TGDTh-4 (bars 1 and 2, respectively) and transformants overexpressing the bipA gene (bars 3 to 7). The error bars indicate standard deviations for three measurements. (B) SDS-PAGE of extracellular culture supernatants. Total proteins (Coomassie blue stained) are shown. Note that α-amylase was directly observed in Coomassie blue-stained gels as it was the most abundant protein. Lane 1, molecular mass markers (molecular masses are indicated on the left); lanes 2 and 3, A. awamori control strains lpr66 and TGDTh-4, respectively; lanes 4 to 8, transformants overexpressing the bipA gene. (C) Immunoblot of secreted glucoamylase levels obtained with anti-glucoamylase antibodies (1:30,000). Lanes 1 and 2, A. awamori control strains lpr66 and TGDTh-4, respectively; lanes 3 to 7, transformants overexpressing the bipA gene. The molecular masses of size markers are indicated on the left.
Attenuation of the bipA gene in A. awamori: some transformants show very high levels of the antisense RNA.
To determine if low BiPA levels might be limiting for thaumatin secretion, downregulation of the bipA gene was carried out by using an antisense RNA strategy. An expression cassette containing a truncated bipA gene in the antisense position under control of the PgdhA promoter (Fig. 5A) was introduced into the thaumatin-producing strain A. awamori TGP-3 (18).
Southern analysis of 10 randomly isolated transformants revealed the existence of several clones with different numbers of copies of the antisense bipA gene (Fig. 5B). A 3.3-kb band corresponding to the endogenous bipA gene was found in both transformant and control strains. Another band at 1.1 kb corresponding to the intact antisense construct was observed only in the transformants. In some strains other bands corresponding to noncanonical integration events were also observed.
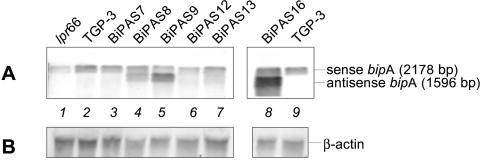
The six transformants that contained only intact copies of the antisense cassette were selected for further study. Northern blot analysis showed that there was a 1,596-bp signal corresponding to antisense bipA RNA in some of the transformants, compared to the 2,178-bp bipA RNA signal observed in both transformants and the TGP-3 and lpr66 control strains (Fig. 6). Higher levels of expression of bipA antisense RNA were observed in transformants BiPAS8, BiPAS9, and BiPAS16. In these transformants, the antisense transcript levels were similar to or higher than (e.g., transformants BiPAS9 and BiPAS16) those of the endogenous bipA gene.
FIG. 6.
(A) Northern blot analysis of transformants containing the bipA antisense cassette. Hybridization was performed with a 1,900-bp BamHI fragment of the bipA gene as the probe. Lane 1, control strain lpr66 (untransformed); lane 2, control strain TGP-3, containing a tha expression cassette; lanes 3 to 9, A. awamori antisense bipA transformants. The upper hybridizing band corresponds to the bipA transcript (2,178 bp). The lower band corresponds to the bipA antisense RNA (1,596 bp). (B) Northern blot analysis of the same samples. A 0.83-kb NcoI-KpnI fragment of the constitutive A. nidulans β-actin gene was used as the probe in control hybridizations.
By comparing of the levels of bipA RNA in the TGP-3 and lpr66 control strains (Fig. 6), we concluded that overproduction of thaumatin in the TGP-3 strain produced a UPR that resulted in overexpression of the bipA chaperone gene in this strain (which showed levels of bipA expression that were 10% higher than those of the lpr66 control strain), as observed also in the TGDTh-4 strain (see above).
RT-PCR amplification confirmed that there were high levels of the antisense RNA.
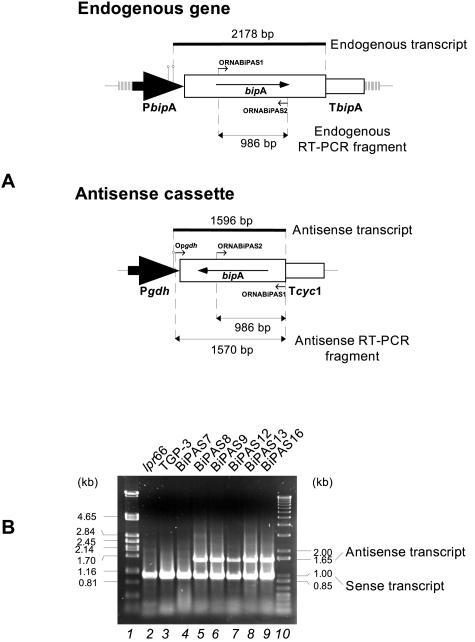
The presence of antisense RNA in transformant strains was also studied by RT-PCR (Fig. 7A). By using purified mRNA as the template, an amplified 986-bp fragment corresponding to the sense transcript of the endogenous bipA gene was observed in both transformant and control strains when ORNABiPAS1 and ORNABiPAS2 were used as the primers. In addition, another amplified fragment, a 1,570-bp fragment corresponding to the antisense bipA RNA, was observed in most of the transformants analyzed but not in the control strains (Fig. 7B). These results confirmed the functionality of the antisense construct.
FIG. 7.
RT-PCR analysis of bipA transcripts in transformants containing the antisense construction. (A) Diagram of the hybridizing fragments from both endogenous and antisense bipA transcripts used in Northern blot analysis (thick lines) and RT-PCR products expected in both cases (thin lines with an arrowhead at each end). The large black arrows represent promoters, and the open boxes with an arrow inside represent bipA sense and antisense genes. The empty open boxes represent terminators. The lollipop symbols indicate transcription start points. The primers used are represented by small arrows. (B) RT-PCR products from mRNA of the lpr66 and TGP-3 control strains (lanes 2 and 3) and antisense transformants (lanes 4 to 9). Lanes 1 and 10 contained size markers (λ PstI and 100-bp ladder). Oligonucleotides ORNABiPAS1, ORNABiPAS2, and Opgdh were used as the primers. The 1,570-bp band corresponds to the bipA sense transcript, and the 986-bp band corresponds to the bipA antisense mRNA.
bipA gene downregulation limits thaumatin secretion: correlation of antisense RNA levels with secreted thaumatin.
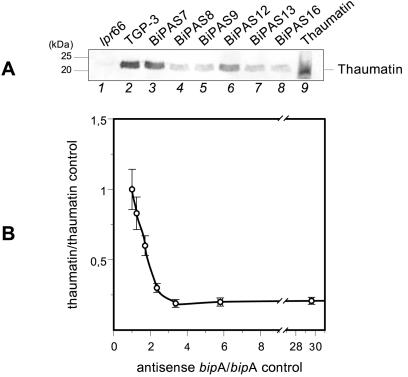
Culture supernatants from transformants containing the antisense RNA cassette were analyzed with anti-thaumatin antibodies (Fig. 8A). The transformants overexpressing the antisense RNA, particularly transformants BiPAS8 and BiPAS9, showed levels of secreted thaumatin that were up to 80% lower than those of control strain TGP-3 (Fig. 8B). On the other hand, transformants BiPAS7 and BiPAS12, which produced low levels of antisense RNA or no antisense RNA, showed minor differences in thaumatin secretion compared to the control strain.
FIG. 8.
(A) Immunoblots of secreted thaumatin in control strains and A. awamori antisense bipA transformants as revealed by anti-thaumatin antibodies (1:15,000). Lanes 1 and 2, A. awamori control strains lpr66 (untransformed) and TGP-3 (containing a tha expression cassette), respectively; lanes 3 to 8, A. awamori BiPAS transformants containing the bipA antisense cassette; lane 9, purified 22-kDa thaumatin from T. daniellii (50 ng). The molecular masses of markers are indicated on the left. (B) Correlation between thaumatin secretion and antisense bipA expression levels of the control strain and the different A. awamori transformants containing the antisense bipA cassette. The data are expressed in units relative to the control strain levels. From left to right the data points correspond to the following transformants: TGP3, BiPAS7, BiPAS12, BiPAS13, BiPAS8, BiPAS9, and BiPAS16.
The quantitative correlation between antisense bipA gene expression and thaumatin secretion (Fig. 8B) showed that an increase in bipA antisense levels caused a decrease in thaumatin secretion up to a certain level of antisense bipA gene expression, above which the secreted levels of thaumatin did not decrease further.
Effect of bipA gene downregulation on secretion of homologous proteins.
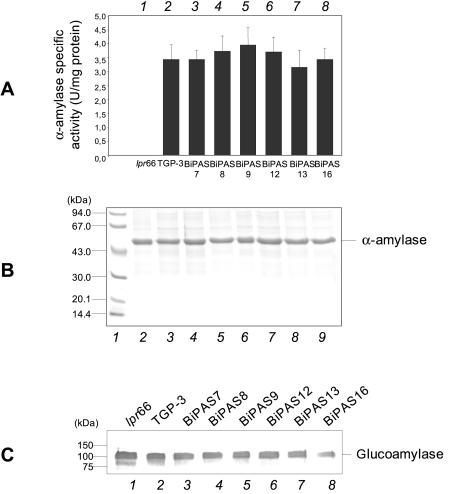
No apparent differences between antisense transformants and control strains were found when the α-amylase activity was quantified (Fig. 9A) or the protein was observed directly in SDS-PAGE gels (Fig. 9B). The levels of glucoamylase, as analyzed by Western blotting, were not different in the transformants and the control strains (Fig. 9C).
FIG. 9.
Secreted homologous proteins α-amylase and glucoamylase in transformants containing the antisense cassette. (A) Extracellular α-amylase activity of A. awamori control strains lpr66 and TGP3 (bars 1 and 2, respectively) and transformants containing the bipA antisense cassette (bars 3 to 9). The error bars indicate standard deviations for three measurements. (B) SDS-PAGE of extracellular total proteins. Note that α-amylase is directly observed in Coomassie blue-stained gels as it is the most abundant protein. Lane 1, molecular mass markers; lanes 2 and 3, A. awamori control strains lpr66 and TGP-3, respectively; lanes 4 to 9, transformants containing bipA antisense cassette. (C) Immunoblots of secreted glucoamylase with anti-glucoamylase antibodies (1:30,000). Lanes 1 and 2, A. awamori control strains lpr66 and TGP-3, respectively; lanes 3 to 7, transformants containing the bipA antisense cassette. The molecular masses of markers are indicated on the left.
In summary, a defined level of bipA gene expression is required for optimal thaumatin secretion, whereas reduction of bipA gene expression by antisense RNA decreased thaumatin secretion. It is interesting that this clear effect on thaumatin production was not observed with the homologous proteins α-amylase and glucoamylase (see Discussion).
DISCUSSION
In filamentous fungi, as in other organism, the BiPA-encoding gene has a basal expression level under normal growth conditions, and it is overexpressed in cellular stress situations, such as glucose starvation, heat shock, and conditions that trigger the UPR (25, 35, 38).
We observed in this work the existence of a clear UPR in A. awamori (increased bipA mRNA levels) following expression of the thaumatin gene (Fig. 2A and 6A). Increases in bipA transcript levels due to overexpression of a recombinant protein have been observed in various yeast and Aspergillus strains (23, 28, 32, 36). The same UPR phenomenon was also observed when we measured another chaperone, PDI, in the same thaumatin-overproducing strains.
The main conclusion of this work is that a defined level of bipA gene expression is required for optimal thaumatin secretion. When this level is reached, the secreted levels of thaumatin increase up to 2.5-fold.
Conversely, downregulation of the bipA gene by antisense RNA in A. awamori causes a large decrease (up to 80%) in the secreted levels of this heterologous protein.
Disruption of the bipA gene seems to be lethal for most organisms (17, 31). Therefore, attenuation of bipA expression by antisense RNA was successful. Decreases in the protein secretion level in different organisms were also observed by other authors when the bipA gene was downregulated (30).
It is interesting that the secreted levels of homologous proteins, such as α-amylase and glucoamylase, do not seem to be affected by these two strategies. These proteins, which are abundantly secreted by A. awamori, appear to have a lower chaperone requirement than the exogenous protein thaumatin. Presumably, secretion of the homologous α-amylase and glucoamylase proteins by A. awamori has adapted to a lower chaperone requirement, whereas plant thaumatin secretion is much more dependent on the BiPA (this study) and PDI (21) levels.
Increases in the secreted levels of other proteins due to overexpression of the bipA gene have been observed in the yeast S. cerevisiae (13, 16, 34). In other cases, no effect on secretion of specific proteins has been observed (6, 7, 15, 17, 30), and even decreases in the secreted levels of the proteins studied have been reported (6, 7, 37).
These different effects on protein secretion caused by overexpression or downregulation of the bipA gene could be due to (i) the multiple cellular functions in which the chaperone BiPA is involved; (ii) the importance of each function in a particular translocation process; and (iii) the different nature of the secreted proteins studied (5). It has also been reported that BiPA is not required for secretion of some specific proteins because they do not interact with BiPA upon transport through the endoplasmic reticulum (22).
Other chaperones, including the foldase PDI (21, 39), may also have a stimulatory effect on heterologous protein secretion (29); the effect of BiPA on thaumatin secretion may increase the effect reported for other chaperones. The increase in the amount of thaumatin produced in bipA-overexpressing transformants might be useful for development of strains of industrial interest.
Acknowledgments
This work was supported by grant 1FD-1111-002-01 from CICYT, Madrid, Spain, and by a generic project of the Agencia de Desarrollo, Junta de Castilla y León, Valladolid, Spain.
We thank D. Archer and D. Jeenes for providing antibodies and B. Martín, J. Merino, A. F. Casenave, and M. Álvarez for their excellent technical assistance.
REFERENCES
- 1.Archer, D. B., and J. F. Peberdy. 1997. The molecular biology of secreted enzyme production by fungi. Crit. Rev. Biotechnol. 17:273-306. [DOI] [PubMed] [Google Scholar]
- 2.Bole, D. G., L. M. Hendershot, and J. F. Kearney. 1986. Posttranslational association on immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 102:1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoza, R. E., F. J. Moralejo, S. Gutiérrez, J. Casqueiro, F. Fierro, and J. F. Martín. 1998. Characterization and nitrogen-source regulation at the transcriptional level of the gdhA gene of Aspergillus awamori encoding an NADP-dependent glutamate dehydrogenase. Curr. Genet. 34:50-59. [DOI] [PubMed] [Google Scholar]
- 4.Cardoza, R. E., S. Gutiérrez, N. Ortega, A. Colina, J. Casqueiro, and J. F. Martín. 2003. Expression of a synthetic copy of the bovine chymosin gene in Aspergillus awamori from constitutive and pH-regulated promoters and secretion using two different pre-pro sequences. Biotechnol. Bioeng. 83:249-259. [DOI] [PubMed] [Google Scholar]
- 5.Conesa, A., P. J. Punt, N. van Luijk, and C. A. M. J. J. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 6.Conesa, A., D. Jeenes, D. B. Archer, C. A. M. J. J. van den Hondel, and P. J. Punt. 2002. Calnexin overexpression increases manganese peroxidase production in Aspergillus niger. Appl. Environ. Microbiol. 68:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorner, A. J., L. C. Wasley, and R. J. Kaufman. 1992. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 11:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faus, I. 2000. Recent developments in the characterization and biotechnological production of sweet-tasting proteins. Appl. Microbiol. Biotechnol. 53:145-151. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez, F. J., S. Gutiérrez, J. Velasco, E. Montenegro, A. T. Marcos, and J. F. Martín. 1994. Molecular characterization of three loss-of-function mutations in the isopenicillin N-acyltransferase gene (penDE) of Penicillium chrysogenum. J. Bacteriol. 176:4941-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-González, M. D., J. F. Martín, T. Vigal, and P. Liras. 1991. Characterization, expression in Streptomyces lividans, and processing of the amylase of Streptomyces griseus IMRU 3570: two different amylases are derived from the same gene by an intracellular processing mechanism. J. Bacteriol. 173:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouka, R. J., P. J. Punt, and C. A. van der Holden. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Hammond, C., and A. Helenius. 1995. Quality control in the secretory pathway. Curr. Opin. Cell Biol. 7:523-529. [DOI] [PubMed] [Google Scholar]
- 13.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 14.Hijarrubia, M. J., J. Casqueiro, S. Gutiérrez, F. J. Fernández, and J. F. Martín. 1997. Characterization of the bip gene of Aspergillus awamori encoding a protein with an HDEL retention signal homologous to the mammalian BiP involved in polypeptide secretion. Curr. Genet. 32:139-146. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, T. A., J. J. Eiden, P. Bourgarel, T. Meo, and M. J. Betenbaugh. 1994. Effects of co-expression chaperone BiP on functional antibody production in the baculovirus system. Protein Expr. Purif. 5:595-603. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M. D., K. C. Han, H. A. Kang, S. K. Rhee, and J. H. Seo. 2003. Coexpression of BiP increased antithrombotic hirudin production in recombinant Saccharomyces cerevisiae. J. Biotechnol. 101:81-87. [DOI] [PubMed] [Google Scholar]
- 17.Li, L.-J., X. Li, A. Ferrario, N. Rucker, E. S. Liu, S. Wong, C. J. Gomer, and A. S. Lee. 1992. Establishment of a Chinese hamster ovary cell line that expresses grp78 antisense transcripts and suppresses A23187 induction of both GRP78 and GRP94. J. Cell. Physiol. 153:575-582. [DOI] [PubMed] [Google Scholar]
- 18.Moralejo, F. J., R. E. Cardoza, S. Gutiérrez, and J. F. Martín. 1999. Thaumatin production in Aspergillus awamori by use of expression cassettes with strong fungal promoters and high gene dosage. Appl. Environ. Microbiol. 65:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moralejo, F. J., R. E. Cardoza, S. Gutiérrez, H. Sisniega, I. Faus, and J. F. Martín. 2000. Overexpression and lack of degradation of thaumatin in an aspergillopepsin A-defective mutant of Aspergillus awamori containing an insertion in the pepA gene. Appl. Microbiol. Biotechnol. 54:772-777. [DOI] [PubMed] [Google Scholar]
- 20.Moralejo, F. J. 2000. Obtención de cepas de Aspergillus awamori modificadas genéticamente para la producción de proteínas heterólogas. Ph.D. thesis. Universidad de León, León, Spain.
- 21.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martín. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 22.Morris, J. A., A. J. Dorner, C. A. Edwards, L. M. Hendershot, and R. J. Kaufman. 1997. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 272:4327-4334. [DOI] [PubMed] [Google Scholar]
- 23.Mulder, H. J., M. Saloheimo, M. Penttilä, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger and up-regulates its own transcription. Mol. Genet. Genomics 271:130-140. [DOI] [PubMed] [Google Scholar]
- 24.Munro, S., and H. R. B. Pelham. 1986. An Hsp70-like protein in the ER: identity with the 78 kd glucose regulated protein and immunoglobulin heavy chain binding protein. Cell 46:291-300. [DOI] [PubMed] [Google Scholar]
- 25.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. M. J. J. van den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedrazzini, E., and A. Vitale. 1996. The binding protein (BiP) and the synthesis of secretory proteins. Plant Physiol. Biochem. 34:207-216. [Google Scholar]
- 27.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 28.Punt, P. J., I. A. van Gemeren, J. Drint-Kuijvenhoven, J. G. Hessing, G. M. van Muijlwijk-Harteveld, A. Beijersbergen, C. T. Verrips, and C. A. M. J. J. van den Hondel. 1998. Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein, in the secretion of homologous and heterologous proteins in black aspergilli. Appl. Microbiol. Biotechnol. 50:447-454. [DOI] [PubMed] [Google Scholar]
- 29.Robinson, A. S., V. Hines, and K. D. Wittrup. 1994. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology 12:381-384. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, A. S., J. A. Bockhaus, A. C. Voegler, and K. D. Wittrup. 1996. Reduction of BiP levels decreases heterologous protein secretion in Saccharomyces cerevisiae. J. Biol. Chem. 271:10017-10022. [DOI] [PubMed] [Google Scholar]
- 31.Rose, M. D., L. M. Misra, and J. P. Vogel. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211-1221. [DOI] [PubMed] [Google Scholar]
- 32.Sagt, C. M., W. H. Muller, J. Boonstra, A. J. Verkleij, and C. T. Verrips. 1998. Impaired secretion of a hydrophilic cutinase by Saccharomyces cerevisiae correlates with an increased association with immunoglobulin heavy-chain binding protein (BiP). Appl. Environ. Microbiol. 64:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Shusta, E. V., R. T. Raines, A. Pluckthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 35.Techel, D., T. Hafker, S. Muschner, M. Reimann, Y. Li, C. Monnerjahn, and L. Rensing. 1998. Molecular analysis of a glucose-regulated gene (grp78) of Neurospora crassa. Biochim. Biophys. Acta 1397:21-26. [DOI] [PubMed] [Google Scholar]
- 36.Valkonen, M., M. Ward, H. Wang, M. Penttilä, and M. Saloheimo. 2003. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 69:6979-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Heide, M., C. P. Hollenberg, I. J. van der Klei, and M. Veenhuis. 2002. Overproduction of BiP affects the secretion of Aspergillus niger glucose oxidase by the yeast Hansenula polimorfa. Appl. Microbiol. Biotechnol. 58:487-494. [DOI] [PubMed] [Google Scholar]
- 38.van Gemeren, I. A., P. J. Punt, A. Drint-Kuyvenhoven, M. P. Broekhuijsen, A. van't Hoog, A. Beijersbergen, C. T. Verrips, and C. A. M. J. J. van den Hondel. 1997. The ER chaperone encoding bipA gene of black aspergilli is induced by heat shock and unfolded proteins. Gene 198:43-52. [DOI] [PubMed] [Google Scholar]
- 39.van Gemeren, I. A., A. Beijersbergen, C. A. M. J. J. van den Hondel, and C. T. Verrips. 1998. Expression and secretion of defined cutinase variants by Aspergillus awamori. Appl. Environ. Microbiol. 64:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witty, M., and J. D. Higginbotham. 1994. Thaumatin. CRC Press, Boca Raton, Fla.
- 41.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]