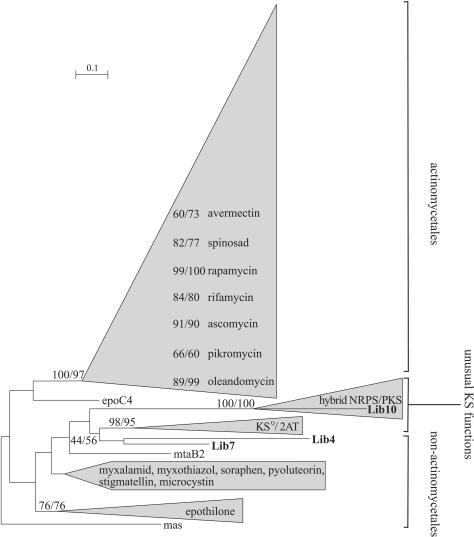
FIG. 1.
Phylogenetic analysis of KS domains. The reconstruction was computed for 207 protein sequences on 413 sites by the ML method (BIONJ, JTT matrix) and the distance method (NJ, PAM matrix) with 500 bootstrap replicates. The group named hybrid NRPS/PKS concerns KS domains preceded by an NRPS. KSQ domains and KS domains from loading modules with the organization ACP-KS-AT-AT-ACP are clustered and are called the KSQ/2AT group. As the tree topologies obtained with both methods are similar, bootstrap values are indicated as ML/NJ. The strongly clustered KS domains of a PKSI are given with the name of the produced polyketide and their respective bootstrap values. The number of amino acid substitutions is proportional to the length of the scale. Sequences obtained from the metagenomic library are given in boldface type. The entire tree is available upon request.