Abstract
A putative G1 cyclin gene, Antma;CycD1;1 (CycD1), from Antirrhinum majus is known to be expressed throughout the cell cycle in the meristem and other actively proliferating cells. To test its role in cell cycle progression, we examined the effect of CycD1 expression in the tobacco (Nicotiana tabacum) cell suspension culture BY-2. Green fluorescent protein:CycD1 is located in the nucleus throughout interphase. Using epitope-tagged CycD1, we show that it interacts in vivo with CDKA, a cyclin dependent protein kinase that acts at both the G1/S and the G2/M boundaries. We examined the effect of induced expression at different stages of the cell cycle. Expression in G0 cells accelerated entry into both S-phase and mitosis, whereas expression during S-phase accelerated entry into mitosis. Consistent with acceleration of both transitions, the CycD1-associated cyclin dependent kinase can phosphorylate both histone H1 and Rb proteins. The expression of cyclinD1 led to the early activation of total CDK activity, consistent with accelerated cell cycle progression. Continuous expression of CycD1 led to moderate increases in growth rate. Therefore, in contrast with animal D cyclins, CycD1 can promote both G0/G1/S and S/G2/M progression. This indicates that D cyclin function may have diverged between plants and animals.
INTRODUCTION
The major control points in eukaryotic cell cycle progression are before DNA synthesis (G1/S boundary) and before the onset of mitosis (G2/M boundary) (Pines, 1995; Huntley and Murray, 1999). Cyclins are rate-limiting proteins that regulate these transitions through activation and targeting of cyclin dependent kinases (CDKs) to specific subcellular compartments and substrates (Jackman et al., 2003). Most organisms contain several different groups of cyclins, which often have specialized functions. For example, in animal cells, B cyclins are implicated in mitotic entry (Minshull et al., 1989), whereas D cyclins play a role in cell cycle reentry in response to extracellular signals (Sherr, 1994), and E cyclins promote entry into S-phase (Geng et al., 2003).
Higher plant genomes also contain different groups of cyclin genes that have been classified according to structural characteristics and named according to the most similar group of mammalian cyclins (Renaudin et al., 1996). However, plant cyclins form phylogenetically distinct groups from mammalian cyclins, suggesting that cyclin function had not finally specialized before the evolutionary divergence of the plant and animal lineages. Moreover, each group of plant cyclins tends to have many more members than the equivalent group in animals. Thus, the D cyclin family is represented by three members in mammals but 10 or more in plants. It is difficult, therefore, to infer function of plant cyclins solely from homology.
Plant D cyclins were originally identified by functional complementation in a yeast strain deficient for G1 cyclins (Soni et al., 1995) and fall into at least four structurally distinct groups.
D cyclins are expressed throughout the cell cycle in actively proliferating plant cells (Sorrell et al., 1999; Gaudin et al., 2000, Menges and Murray, 2002). Their expression can be modulated by growth signals (Soni et al., 1995; Fuerst et al., 1996; Riou-Khamlichi et al., 1999, 2000; Sorrell et al., 1999; Gaudin et al., 2000) in a manner somewhat analogous to mammalian D cyclins and extracellular growth factors (Motokura and Arnold, 1993; Sewing et al., 1993). In animals, D cyclins are transcribed in response to extracellular growth signals and they associate with CDKs to produce an active Rb protein kinase that phosphorylates the Rb protein, resulting in the release and activation of E2F transcription factors that are responsible for the transcription of S-phase genes (reviewed in Sherr and Roberts, 1999; Shen, 2002). The identification of plant proteins closely related to animal Rb suggests that a similar regulatory mechanism exists in plants. Plant D cyclins also contain a functional Rb binding motif, and Rb is a possible substrate (Ach et al., 1997; Huntley et al., 1998; Nakagami et al., 1999, 2002).
In Arabidopsis thaliana, there are at least 10 D cyclin genes forming several structurally distinct groups (Arabidopsis Genome Initiative, 2000; Vandepoele et al., 2002). There is likely to be extensive functional redundancy because the genome-wide insertional mutagenesis surveys have yet to report phenotypes for D cyclin knockouts (Campisi et al., 1999, Swaminathan et al., 2000). The Arath;CycD1;1 mutant plants have no obvious phenotype (O. Koroleva and J. Doonan, unpublished data). However, overexpression of selected D cyclin genes indicates that two of the three main groups have different functions. Thus, Arath;CycD2;1 appears to be rate limiting for progression through G1 in roots and shoots, leading to faster growth rate of transgenic tobacco (Nicotiana tabacum) plants (Cockcroft et al., 2000), whereas overexpression of Arath;CycD3;1 promotes prolonged cell proliferation in leaves and inhibits differentiation (Dewitte et al., 2003). Ectopic expression of Arath;CycD3;1 in endoreduplicating trichome cells induced cell division (Schnittger et al., 2002).
However, constitutive expression of positive cell cycle regulators can be difficult to interpret because an effect in one phase of the cycle may be masked by compensatory changes in another. Cooper (1998) argued that it is not possible to distinguish the phase-specific effects of G1 cyclin expression without considering changes in the overall cellular growth rate because an observed shortening of the G1 phase could be because of an increase in the rate of mass synthesis in all phases of the cell cycle. Here, we have used an experimental approach that allowed us to solve this logical problem by inducing D1 cyclin expression at specific times of the cell cycle and therefore temporally dissecting the function of D1 cyclin during G1/S and S/G2 phases of the cell cycle.
We have used a modified AlcR/AlcA two-component gene expression system that permits inducible gene expression in cell cultures in response to added dexamethasone (Dex) instead of ethanol used for the original AlcR/AlcA system (Roslan et al., 2001) to monitor the consequences of D1 cyclin gene expression. The only data available on functional analysis of D1 cyclins in plants relates to their mRNA expression patterns in Arabidopsis (Soni et al., 1995), Antirrhinum majus (Gaudin et al., 2000), and Jerusalem artichoke (Helianthus tuberosus) (Freeman et al., 2003) where these genes are expressed in proliferating tissues. In Antirrhinum plants, cyclin Antma;CycD1;1 (CycD1) transcripts are present in meristems, but expression is continuous throughout the cell cycle (Gaudin et al., 2000). Here, we show that CycD1 is primarily localized to the nucleus and that it interacts with CDKA, a cyclin dependent protein kinase that is known to stimulate both G1/S and G2/M transition. Our data indicate that cyclin D1 can act at multiple points in the cell cycle to promote progression unlike mammalian D cyclins (whose primary function is implemented at G1) and is not simply limited to the G1/S boundary. Induction of expression of CycD1 led to elevated activity of both total CDK fraction and CycD1 interacting kinase throughout all stages of the cell cycle with prominent peaks associated with S-phase and mitosis. Expression of CycD1 in G0 cells accelerates entry into S-phase as would be predicted for G1 cyclin. However, expression of CycD1 during or after S-phase also led to faster progression through G2 into mitosis.
RESULTS
CycD1 Is Located in the Interphase Nucleus
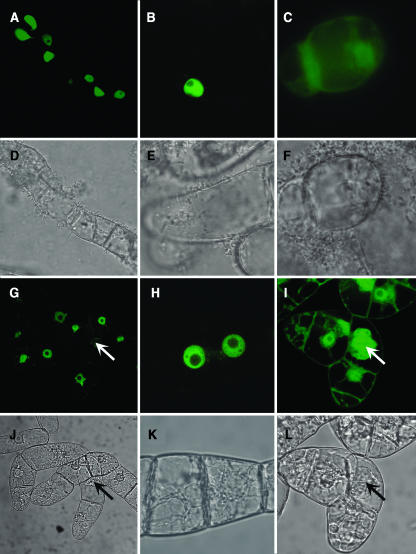
The location and timing of a cyclin's appearance can provide clues as to its probable function (Draviam et al., 2001; Jackman et al., 2003). Therefore, as a first step, we transiently expressed green fluorescent protein (GFP):CycD1 under the 35S promoter of Cauliflower mosaic virus (CaMV35S) promoter in the Arabidopsis cell suspension culture. The GFP:CycD1 fusion protein localized specifically to the nucleus, usually with less signal in nucleoli (Figures 1A, 1B, 1D, and 1E). Control experiments using soluble GFP under the CaMV35S promoter gave strong fluorescence in both cytosol and nuclei (Figures 1C and 1F). We were unable to find any mitotic cells containing a detectable GFP:CycD1 in our transient experiments, so to study the dynamics of localization during the cell cycle, we made stable transgenic lines in BY-2 expressing GFP:CycD1. Several independent lines were produced, all of which showed the same pattern of nuclear localization as in Arabidopsis, albeit at a much lower level. We also observed some small fluorescent bodies surrounding the nuclei, but these were also found in control nontransformed cells imaged under the same conditions (data not shown). BY-2 cell suspensions have a relatively high mitotic index compared with Arabidopsis, and we were able to identify mitotic cells readily (Figures 1G, 1I, 1J, and 1L). The GFP:CycD1 signal associated with the chromatin in mitotic cells is dramatically reduced compared with that in the interphase nuclei of near neighbors (Figure 1G). Soluble GFP remains in the spindle domain of mitotic cells in the GFP control line (Figure 1I). The persistence of GFP:CycD1 in the nuclear domain throughout interphase suggested to us that CycD1 may play additional roles during the cell cycle, in addition to its presumed role during G1.
Figure 1.
Localization of GFP:CycD1 Fusion Protein and GFP in Living Cells.
(A) to (F) Transiently expressed in Arabidopsis cell culture.
(A) and (B) Confocal images of GFP fluorescence in cells transformed by GFP-CycD1;.
(C) and (F) Confocal image of soluble GFP fluorescence and corresponding phase contrast image.
(D) and (E) Corresponding phase contrast images.
(G) to (L) Stable expression in BY-2 cell culture. Arrows indicate mitotic cells.
(G) and (H) Confocal images of GFP fluorescence in cells transformed by GFP-CycD1;.
(I) and (L) Confocal image of soluble GFP fluorescence and corresponding phase contrast image.
(J) and (K) Corresponding phase contrast images.
Dex-Induced Expression of CycD1
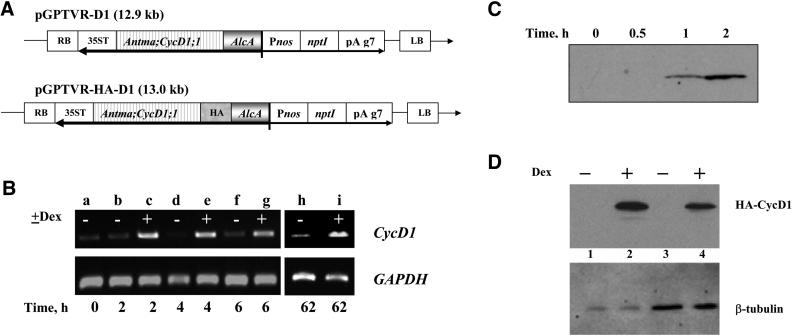
To define the role of CycD1 during the cell cycle, we exploited the AlcR:GR/AlcA gene expression system, a chemically inducible gene switch modified for use in cell suspensions to make it responsive to Dex rather than ethanol. This approach allowed us compare the same cell line with or without CycD1 expression, thus avoiding the problem of variability between transgenic cell lines and to drive CycD1 expression at defined times during the cell cycle. We isolated 16 independent cell lines secondarily transformed with AlcA:D1 or AlcA:HA:D1 expression constructs (Figure 2A) in three different primary lines carrying the modified AlcR:GR transcription factor. After induction with Dex, all AlcA:D1 and AlcA:HA:D1 lines had a significant amount of CycD1 transcript, as judged by RT-PCR. No PCR product was obtained from any primary AlcR:GR lines (data not shown), confirming that D1 primers were specific for the CycD1 and that endogenous tobacco cyclin sequences were not amplified. A time course of CycD1 induction for one of these AlcA:D1 lines with or without induction by 10 μM Dex is shown in Figure 2B. A very low level of expression was observed before induction and expression increased dramatically 2 h after induction, reaching a maximum by 4 h. An elevated level of CycD1 was present in the induced culture for at least 62 h (Figure 2B). Similarly, all 16 secondary transgenic lines had low expression that could be increased by addition of Dex. Protein gel blot analysis of HA-tagged CycD1 lines demonstrated that the protein was first detectable 1 h after induction by 10 μM Dex (Figure 2C), and significant amounts of CycD1 were present 24 h after induction by Dex, whereas in the control culture without induction the protein was undetectable (Figure 2D).
Figure 2.
Inducible Expression of CycD1 in BY-2 Cells.
(A) Inducible cassettes used for transformation. The T-DNA regions of pGPTVR-D1 and pGPTVR-HA-D1 are illustrated. LB, left border; RB, right border.
(B) Time course of CycD1 mRNA expression monitored by RT-PCR, compared with constitutive expression of GAPDH used as loading control. A stationary culture was simultaneously diluted by fresh media and induced (except the control flask) by adding Dex (time = 0). Lane a, culture at time zero; lanes b, d, f, and h, noninduced culture (marked by “−”); lanes c, e, g, and i, induced culture (marked by “+”). Time after induction is shown in hours.
(C) Time course of HA:CycD1 protein expression assessed by protein gel blot with anti-HA antibody. A 4-d-old culture was induced by 10 μM Dex at time 0, and samples were taken as indicated. Equal amounts of protein from induced samples for each time point were loaded on a gel and blotted.
(D) Protein gel blot with anti-HA antibody for two transgenic lines induced by 10 μM Dex for 24 h. Equal amounts were loaded for pairs of induced/noninduced samples, as verified by subsequent blotting with anti β-tubulin antibody. Lane 1, line K 2.3 control; lane 2, line K 2.3 + Dex; lane 3, line K7 control; lane 4, line K7 + Dex.
CycD1 Is an Unstable Protein That Interacts with CDKA in Vivo
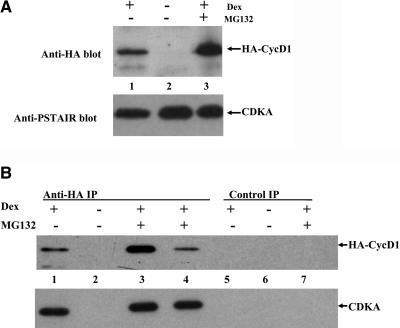
Even after 24 h of induced expression in cultures, the protein extracts contained a relatively low amount of CycD1 protein, consistent with the not very high intensity of GFP:CycD1 fluorescence. We therefore examined the predicted amino acid sequence for motifs involved in protein turnover using the PESTFIND software program and identified two high scoring PEST sequences (potential proteolytic cleavage sites), the most prominent located at the C-terminal part scoring +10 (residues 290 to 309) and another PEST sequence with positive score of +4 located at the N-terminal part (residues 1 to 55). Animal and plant D cyclins contain PEST sequences, regions rich in four residues (Pro, Glu, Ser, and Thr), which are characteristic of many proteins that are rapidly turned over (Rogers et al., 1986). All plant D cyclins isolated to date contain PEST regions, suggesting that they are labile proteins degraded by a similar pathway to animal D-type cyclins (Soni et al., 1995; Oakenfull et al., 2002). To test if CycD1 was subject to proteosome-mediated degradation in cycling cells, we made protein extracts from cultures grown in the presence or absence of the proteosome inhibitor MG132. Those cultures induced by Dex for 24 h and treated with MG132 for 2 h before protein extraction accumulated higher amounts of expressed HA:D1 protein compared with Dex-induced cultures not treated with the proteasome inhibitor (Figure 3A). In noninduced cultures, HA:D1 was not detectable on protein gel blots; therefore, these cells contained a negligible amount of the protein (Figures 2D and 3A). The amount of native CDKA protein, as detected by anti-PSTAIR antibodies, was unaffected by these treatments (Figure 3A).
Figure 3.
Protein Gel Blots of Whole-Cell Extracts and Immunoprecipitated Proteins, Demonstrating Interaction of HA:CycD1 and PSTAIR CDK.
(A) Protein gel blot of whole protein extracts probed with antiserum against HA or PSTAIR motif of CDKA kinases. Lanes 1, line K2.3 + Dex; lane 2, K2.3 control; lane 3, K2.3 + Dex + MG132.
(B) Protein gel blots of anti-HA and control anti-goat IG immunoprecipitates probed with antiserum against HA and against PSTAIR motif of CDKA kinases. Lanes with HA beads: lane 1, line K2.3 + Dex; lane 2, K2.3 control; lane 3, K2.3 + Dex + MG132; lane 4, line K7 + Dex + MG132. Lanes with control IgG beads: lane 5, K2.3 + Dex; lane 6, K2.3 control; lane 7, K2.3 + Dex + MG132.
The in vivo partners of D1 cyclins are unknown. As a first step in determining these, we established conditions for immunoprecipitating HA-tagged CycD1. Anti-HA antibodies attached to agarose beads were used to isolate the tagged protein from total soluble protein extracts made with induced or noninduced cells. The immunoprecipitated proteins were blotted with both anti-HA and anti-PSTAIR antibodies (Figure 3B), clearly demonstrating that A-type CDK had coprecipitated with CycD1. Moreover, CDKA only coprecipitated from protein extracts of cultures induced by Dex (lanes 1, 3, and 4) but not from noninduced culture (lane 2) or nontransformed lines (data not shown), indicating that the immunoprecipitation of CDKA was dependent on the presence of HA:CycD1. No CDKA proteins were precipitated on control anti-goat IgG beads (lanes 5 to 7), further confirming specificity of coprecipitation of CDKA with HA:CycD1 protein. To assess whether CycD1 bound to other CDKs, we probed immunoprecipitates with antibodies against CDKB1;1 (kind gift of Masaki Sekine). The antibody recognized CDKB1;1 protein in whole cell extracts (Figure 4D) but not in fractions immunoprecipitated on anti-HA beads (data not shown).
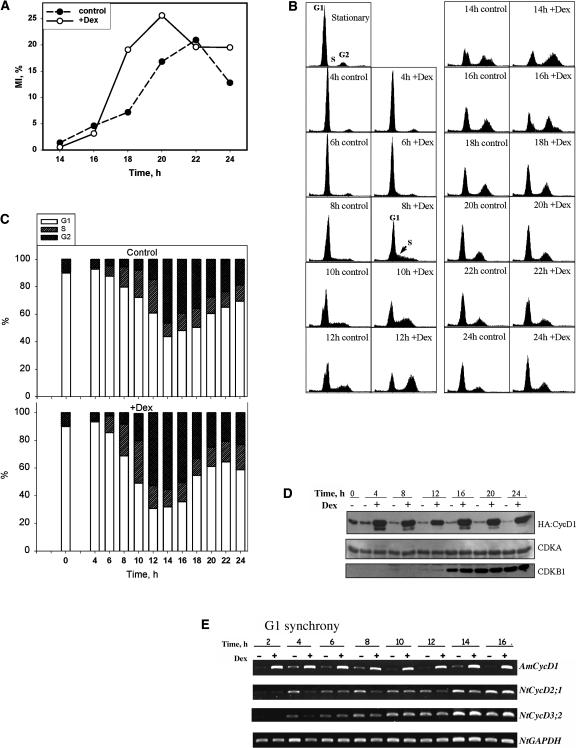
Figure 4.
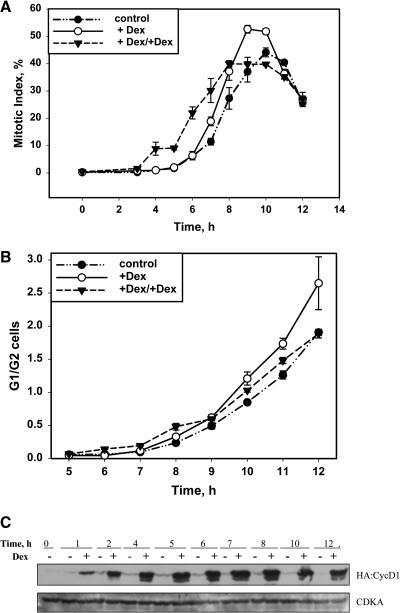
Effect of Expressing CycD1 from G1.
A 10-d-old stationary culture was simultaneously diluted by fresh media and induced by adding 10 μM Dex at time 0.
(A) Time course of MI after dilution with or without induction.
(B) Flow cytometric analysis of cell ploidy, after dilution with (+Dex) or without (control) induction. The positions of the G1 and G2 peaks and S-phase are indicated. Time is indicated in hours after dilution/induction.
(C) DNA histogram of data from (B) showing proportions of G1, S, and G2 cells at different times after dilution with or without induction.
(D) Protein gel blots of whole-cell extracts taken from induced (+) or noninduced cultures (−) in G1 synchrony experiment and probed by antiserum against HA, PSTAIR, or CDKB1.
(E) Expression of endogenous cyclins has not been significantly affected by induction of expression of CycD1, as analyzed by RT-PCR. Total cDNA obtained by reverse transcription of mRNA extracted using magnetic beads from cell extracts was used as a template for amplification of fragments of Antma;CycD1;1, NtCycD2;1, NtCycD3;2, and NtGAPDH as constitutive expression control.
CycD1 Expression during G1 Advances Both S-Phase and Mitotic Entry
To test the effect of CycD1 on overall cell cycle progression, we first examined the effect of continuous CycD1 expression on cells as they were released from stationary phase. Stationary phase cultures were obtained by allowing cultures to age until the mitotic index dropped to undetectable levels (<0.1%). This takes ∼9 to 11 d after subculture, and the cells arrest in G0 with unreplicated DNA (Figure 4B). Such cultures can be induced to reenter the cell cycle by addition of fresh media and they proceed in a semisynchronous manner through G0/G1 transition and G1 into S-phase and mitosis (Figures 4A to 4C). We assayed the behavior of 16 independently isolated cell lines. At 8 h after induction, 8 out of 8 CycD1 lines and 4 out of 8 HA:CycD1 lines had a remarkable increase of cells in S and G2 phases compared with noninduced controls. This marked acceleration of entry into S-phase caused by expression of CycD1 represents an ∼2 h advance and therefore could fully account for the accelerated progression to mitosis. However, this does not rule out the possibility that S and G2 phases were also affected. Several lines were selected for more detailed analysis. If CycD1 is induced at the same time as cells are subcultured into fresh media, entry into mitosis is accelerated as judged by mitotic index (MI) measurements (Figure 4A represents typical experiment) by at least 2 h. CycD1 induction led to a significantly increased MI by 18 h after subculture, when the induced culture had MI of 19% compared with the noninduced culture's 7% (Figure 4A). The induced culture entered mitosis and reached its peak value ∼2 h earlier and also achieved a higher peak value of MI compared with the control (Figure 4A). Thus, expression of CycD1 significantly accelerated cell cycle progression.
To investigate the basis for CycD1-mediated cell cycle acceleration, we followed the dynamics of S-phase entry with or without expression of CycD1. As indicated by flow cytometric analysis (Figures 4B and 4C), 90% of nuclei in stationary BY-2 cells (10 d after subculture) had unreplicated DNA, indicating that they exit the cell cycle mainly from G1 (Planchais et al., 1997). At 8 h after dilution and induction, the induced culture had a significantly higher proportion of cells in S-phase (23%) compared with noninduced culture (15%). This excess of S-phase cells was clearly visible as a shoulder on the G1 peak (Figure 4B). By 12 h, the proportion of nuclei in the G2 peak (53%) exceeded the number in the G1 peak (31%) in induced culture, whereas G1 nuclei (61%) still predominated in the noninduced culture and only 15% contributed to the G2 peak. The highest proportion of cells in S-phase was observed at 10 h after induction/dilution (31%) compared with 21% in noninduced culture. By 24 h after dilution, the proportion of cells in S-phase and the G2/G1 ratio was higher in the induced culture, indicating that the induced culture has progressed faster into the S-phase of the second cell cycle than did the control culture (Figures 4B and 4C). To examine whether the induction of expression of CycD1 has a long-term effect on cell cycle progression, we estimated MI 62 h after induction and found that the induced culture had MI of 7.5%, which was slightly higher than 5.6% in the control culture.
Protein Levels of Overexpressed HA-CycD1, CDKA, and CDKB1 Show Distinct Profiles in G1 Synchronized Culture
We used protein gel blot analysis of whole-cell extracts to follow changes in levels of HA:CycD1 protein, CDKA, and the mitotic marker protein CDKB1 during the cell cycle under induced and noninduced conditions. A transformed line with a high level of inducible expression of HA:CycD1 protein had been chosen for analysis of protein abundance throughout the cell cycle in cells reentering G1 phase or synchronized by aphidicolin. Protein gel blots with anti-HA antibody revealed that a high level of HA:D1 protein was present in induced cultures, whereas noninduced cultures contained only a small amount (Figure 4D). Blotting with anti-PSTAIR antibody showed that CDKA remained at a constant level during the cell cycle as was reported previously by Sorrell et al. (2001). CDKB1 protein was undetectable in stationary cells. After reentering the G1 phase, CDKB1 could be first detected at 12 h (corresponding to late S-phase in G1 synchrony) and then its levels increased gradually, but were slightly higher in induced cultures at 12 and 16 h points (Figure 4D).
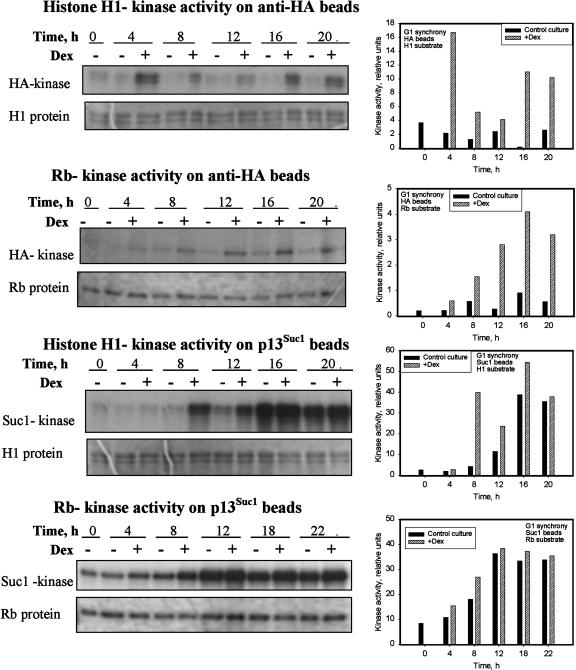
CycD1 Associated Kinase Can Phosphorylate Both Histone H1 and Rb Proteins
To test the substrate specificity of the CycD1-associated kinase at different phases of the cell cycle during the G1 synchrony experiment, we assayed HA-associated kinase activity toward histone H1 and Rb proteins. Immunoprecipitation of HA-tagged cyclin D1 on anti-HA beads allowed us to specifically measure kinase activity of overexpressed CycD1 complex and compare with the endogenous tobacco CDKs purified on p13Suc1 beads. Expression of HA:CycD1 led to an initial increase in HA-associated histone H1 kinase activity detected at the 4 h time point (Figure 5), followed by decline at 8 to 12 h and second peak of activity at 16 to 20 h, which corresponded to the time of mitosis. The noninduced control culture had a very low background level of HA-associated histone H1 kinase activity that did not change significantly during the time course. HA-dependent phosphorylation of Rb protein increased steadily from 4 h (G1) to 16 h (mitosis) in the induced culture (Figure 5). The Rb kinase activity in noninduced culture was low but increased slowly with time.
Figure 5.
Kinase Activity Precipitated on Anti-HA or p13Suc1 Beads during Cell Cycle in a G1 Synchrony Experiment Was Increased upon Expression of HA:CycD1 Either with Histone H1 or Rb Protein as a Substrate.
Autoradiographs with corresponding images of stained gels and quantitative analysis of band optical density are shown for each bead/substrate combination. Expression of HA:CycD1 led to increase in kinase activity precipitated either on anti-HA or p13Suc1 beads, with distinct cell cycle profiles of histone H1 and Rb kinase activity.
CycD1 Expression during G1 Synchrony Increased Total CDK Activity Precipitated on p13Suc1 Beads
To see whether ectopic expression of CycD1 led to changes in overall CDK activity toward either histone H1 or Rb protein, we measured total kinase activity profiles for both these substrates. Precipitation of total CDK activity on p13Suc1 beads brought an order of magnitude higher phosphorylation activity of both histone H1 and Rb proteins, compared with the CycD1-interacting fraction described above. Histone H1 kinase activity in the induced culture was dramatically increased compared with the control at 8, 12, and 16 h (Figure 5). Although after 8 h activity has increased in both, the induced culture still had greater activity than control at 12 and 16 h (mitosis). The noninduced culture showed a continuous increase in kinase activity from G1 through S-phase into mitosis. Kinase activity toward Rb protein followed a very similar trend of steady increase through G1 and S-phase (4 to 12 h) and then a slight decrease at the time of mitosis (18 to 22 h) in both induced and noninduced cultures. However, Rb kinase activity was higher in the induced culture at each time point (Figure 5).
Expression of Endogenous Cyclins in Transformed Lines
To investigate whether ectopic expression of CycD1 influenced the expression profile of the endogenous tobacco cyclins, we have performed RT-PCR amplification of fragments of selected cyclins during G1 and aphidicolin synchrony experiments. Expression of Nicta;CycD2;1 and Nicta;CycD3;2 gradually increased through the S and G2 phase toward mitosis (Figure 4E), whereas Nicta;CycD3;1 expression was very low (data not shown). No differences between induced and noninduced cultures were found for Nicta;CycD3;2 (Figure 4E); the Nicta;CycD2;1 transcripts were slightly less abundant in induced culture at 4, 8, and 12 h time points. However, there was no difference for other time points. We have observed such differences in independent experiments, and they may be related to the stage of the cell cycle. One possibility is that they represent endogenous feedback control of D cyclin transcript levels at particular stages of the cell cycle. No discernible differences were found in transcript abundance of Nicta;CDKA;4, Nicta;CDKB1;1, Nicta;CycA3;2; Nicta;CycB1;1, Nicta;CycD2;1, Nicta;CycD3;1, Nicta;CycD3;2, and Nicta;GAPDH (glyceraldehyde 3-phosphate dehydrogenase) between induced and noninduced cultures at 1, 8, and 12 h after release from aphidicolin, though there were apparent differences in expression levels between these genes and changes for the same genes between different time points (data not shown).
CycD1 Expression during S-Phase Accelerates Entry into Mitosis
Having found that the expression of CycD1 drove faster progression from G0 through G1 phase to S-phase and into the following mitosis, we asked the question whether CycD1 expression could accelerate the cell cycle progression in cells that had already passed the G1/S boundary. If cell cycle acceleration was specific to G1 phase only, then CycD1 expression during or after S-phase should not accelerate cell cycle progression. Aphidicolin, an inhibitor of activity of DNA polymerases α and δ (Sala et al., 1983; Nagata et al., 1992) reversibly arrests cells in early S-phase. We tested the effects of continuous CycD1 induction either from early S-phase or from late S-phase in cell cultures synchronized by aphidicolin. For early S-phase induction, we added Dex together with aphidicolin and then again after the removal of aphidicolin to keep a constant concentration in the media (referred to as the Dex/Dex culture). For late S-phase induction, we added Dex immediately after removal of aphidicolin (referred to as the Dex culture). Noninduced cultures were kept as controls.
The Dex cultures (induced on release from the aphidicolin block) progressed faster into mitosis and achieved a higher maximal value of MI than the control (Figure 6A). The number of mitotic cells by 7 to 10 h was significantly increased, and we estimated that induction of CycD1 expression at this stage accelerated entrance into mitosis by approximately 1 h (Figure 6 represents a typical experiment), suggesting an effect in late S-phase or the G2/M transition. To follow progression into and completion of mitosis, we analyzed appearance of newly divided cells (those with unreplicated DNA or G1) by flow cytometry. The flow cytometric analysis is presented as the ratio between populations of cells in G1 and G2 phases, each calculated as an integral number of cells with DNA content of 2C or 4C in a sample (Figure 6B). The G1/G2 ratio was almost zero at 5 h after the aphidicolin removal, indicating that the vast majority of cells were in G2 phase, and later it gradually increased, indicating appearance of freshly divided G1 cells. By 8 to 9 h after release, the number of cells that passed through mitosis as judged by G1/G2 index (Figure 6B) became significantly higher in cultures induced by Dex, and the difference was maintained during the following several hours. This could be because of the shortening of either S or G2 phase because the earliest time of appearance of HA:CycD1 protein is detected 1 h after induction (Figure 2C). This time point corresponds to the peak in DNA synthesis, and 2 h later the S-phase is practically completed (Sorrell et al., 1999).
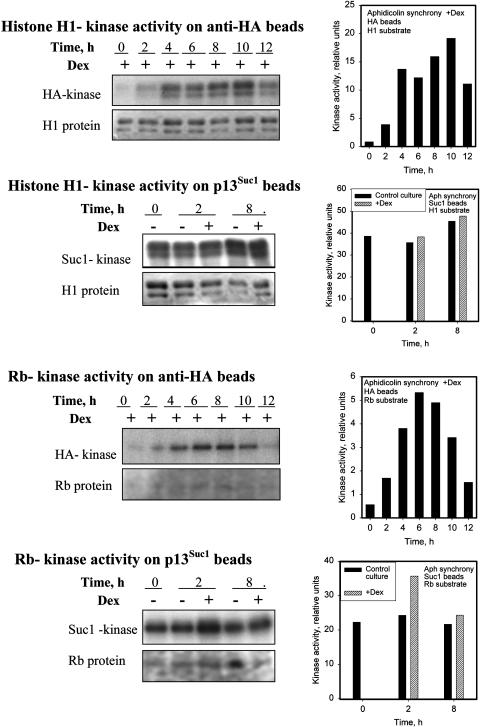
Figure 6.
Effect of CycD1 during Early or Late S-Phase.
A 7-d-old culture was simultaneously diluted by fresh media and arrested in S-phase by 5 μg/mL of aphidicolin for 24 h before release at time 0. Control, received no induction before or after release of aphidicolin block; +Dex, induced with 10 μM Dex after release from the aphidicolin block; +Dex/+Dex, induced twice, upon application and release of aphidicolin block. Each point is an average value ±se representing three independent biological replicas for each treatment.
(A) Time course of MI in induced and control cells.
(B) Time course of G1/G2 nuclei ratio calculated from flow cytometry data.
(C) Protein gel blots of whole-cell extracts taken from induced (+) or noninduced cultures (−) in an aphidicolin synchrony experiment and probed by antiserum against HA and PSTAIR.
Earlier induction (by inducing both during and after the aphidicolin block [Dex/Dex culture]) produced an even greater acceleration in mitosis. A significant number of cells were in mitosis at 4 h after the aphidicolin block was released (Figure 6A), indicating that progression into mitosis was faster with either mode of induction. The initially higher speed of cell cycle progression in the Dex/Dex cultures was most likely achieved as a result of accelerated progression through S-phase. However, 9 h after block release, the number of cells that had passed through mitosis, as judged by G1/G2 index (Figure 6B), became equal in cultures induced by Dex once or twice. By 12 h, the G1/G2 index in the Dex/Dex cultures was lower than in the Dex cultures, indicating that cultures induced in early S-phase entered mitosis earlier but spent a prolonged time in mitosis. It is possible that induction of CycD1 during an aphidicolin-induced S-phase arrest led to damage that then triggered a delay during mitosis. However, we could not find any significant increase in cytological abnormalities during mitosis when we compared Dex/Dex cells to Dex cells or aphidicolin treated controls.
CDKA Shows Constant Protein Level throughout Cell Cycle in Both Induced and Noninduced Aphidicolin-Synchronized Cultures
In cells synchronized by aphidicolin, protein gel blot analysis with anti-HA antibody showed that a high level of HA:D1 protein was present in induced cultures, whereas noninduced cultures contained only a small amount of the expressed protein (Figure 6C). CDKA remained at a constant level during the cell cycle progression from S-phase into mitosis, as was demonstrated by blotting with anti-PSTAIR antibody (Figure 6C), similar to the G1 synchrony experiment described above and consistent with previously reported data by Sorrell et al. (2001).
Increased Kinase Activity with Distinctively Different Time Course for Histone H1 and Rb Proteins Was Caused by CycD1 Expression during Aphidicolin Synchrony
Total CDK kinase activity (precipitated on p13Suc1 beads) in the aphidicolin synchrony experiment was compared between induced and noninduced cultures at two time points: 2 h (S-phase) and 8 h (mitosis). We found that in induced cultures, the kinase activity was always higher; however, this increase represented a small fraction at all time points except in S-phase, where the induced culture had 30% higher Rb phosphorylation activity (Figure 7). The profile of CycD1-interacting CDK activity immunoprecipitated on anti-HA beads had been similar to that observed in the G1 synchrony. Two peaks of activity, the first 4 h after Dex induction and the second in mitosis (8 to 10 h), were found toward the histone H1 substrate as well as a continuous rise of Rb kinase activity until mitotic entry (6 to 8h).
Figure 7.
Kinase Activity Precipitated on Anti-HA or p13Suc1 Beads during Cell Cycle in an Aphidicolin Synchrony Experiment, Either with Histone H1 or Rb Protein as Substrate.
Expression of HA:CycD1 led to moderate increase in kinase activity precipitated either on anti-HA or p13Suc1 beads; characteristically distinct cell cycle profiles of histone H1 and Rb kinase activity immunoprecipitated on anti-HA beads were obtained for cultures expressing HA:CycD1.
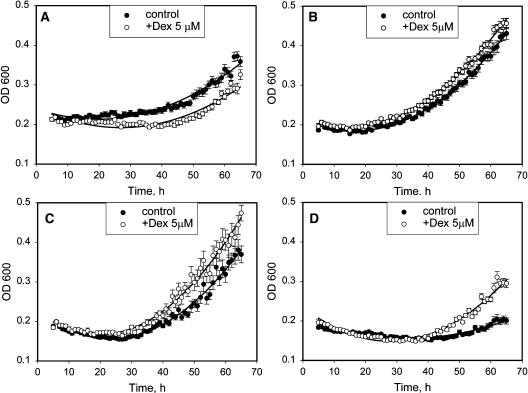
Biomass Accumulation in Transgenic Lines
Because cyclin D expression accelerates cell cycle progression at two key stages of the cell cycle, we predicted that overall growth should be accelerated. Compensation should not occur because there is an acceleration of both G1 and G2 phases. To test this, we followed the change in cell growth rate and overall biomass accumulation using Bioscreen C plate reader shaker incubator. The optical density (OD600) readings provided by Bioscreen C are widely used to follow growth in microbial cultures (Augustin et al., 1999). We have counted cell numbers in BY-2 cultures before and after growing them using Bioscreen C and found that OD600 readings have a strong positive correlation with the cell numbers in samples (data not shown). Induction of CycD1 expression led to an acceleration of growth in the cultures, as compared with the same cultures without induction (Figures 8B to 8D). No significant differences were found for the mean cell volumes between treatments, once again supporting the idea that the increased growth rate is primarily attributable to faster cell cycles. Induction of primary lines carrying the receptor switch AlcR:GR has an inhibitory effect on culture growth (Figure 8A). Because induction produced significant growth rate increases in both the CycD1 and HA:CycD1 lines, the overall effect must have been more dramatic to overcome the small negative effect of the switch.
Figure 8.
Growth of Transgenic BY-2 Cell Lines Monitored with Bioscreen C, with or without Induction by 5 μM Dex.
Each point on the graphs is an average value ±se of OD600 for 10 separate wells, each containing 200 μL of BY-2 suspension culture.
(A) A primary AlcR:GR line, containing only the AlcR:GR transactivator.
(B) A secondary line, containing an AlcA:CycD1 expression cassette.
(C) and (D) Two independent secondary lines, containing an AlcA:HA:CycD1 expression cassette.
DISCUSSION
In this report, we show that CycD1, an unstable CDKA interacting protein localized to the nucleus during interphase, has the potential to accelerate cell cycle progression during at least two discrete phases of the cell cycle.
CycD1 Location, Stability, and Interaction with CDKA
Coimmunoprecipitation of HA-tagged CycD1 with an A-type CDK containing a PSTAIRE motif is consistent with the known interactions of other D-type cyclins (reviewed in Oakenfull et al., 2002). Arath;CycD2;1 and Arath;CycD3;1 have been previously shown to interact with CDKA in vivo (Healy et al., 2001). The interaction between Arath;CycD1;1 and CDKA has been demonstrated by in vitro pull-down assays (Kono et al., 2003), and our study confirms that this interaction can occur in vivo. The GFP localization study lends indirect support to this idea. Nuclear localization is consistent with what is known about other D cyclins in plants; Nicta;CycD3;3 was found primarily in the nucleus (Nakagami et al., 2002). We have previously shown that CDKA:GFP fusions are located to the nuclei during interphase (Weingartner et al., 2001) in a very similar pattern to that of GFP:CycD1. However, whereas CDKA:GFP transfers from the chromatin domain to the microtubules at metaphase, GFP:CycD1 either disperses or is degraded during early mitosis (Figure 1G). CycD1 protein levels are enhanced by treatment with MG132, a specific proteasome inhibitor (Figure 3A), suggesting that protein levels are controlled, at least in part, by proteasome-mediated degradation. We were unable to find evidence that cyclin D1 interacts with CDKB1, a kinase closely related to CDKA, so we conclude that nuclear located CycD1 interacts in vivo primarily with CDKA, a kinase that can act at multiple stages of the cell cycle. This interaction led to formation of functionally active histone H1 and Rb kinase that may be further regulated by posttranslational modification at S and M phase. This kinase activity was partly inactivated during M phase (Figure 7), correlating with the disappearance of GFP-CycD1 from the nucleus (Figure 1). Whether this is because of specific protein degradation remains to be determined.
CycD1 Accelerates Progression through the Cell Cycle in BY-2 Cells
The induction of CycD1 at defined points during the cell cycle allowed us to dissect its role during G0/G1/S and S/G2/M phases. This directly addresses the logical problem that constitutive expression of a cyclin could lead to changes in the overall cellular growth rate and not necessarily only to shortening of G1 phase (Cooper, 1998). Therefore, we conclude that overexpression of CycD1 can affect not only the length of G0/G1 but also can directly affect S/G2 phases. Our results clearly indicate that, unlike mammalian D cyclins, CycD1 functions not only at the G0/G1/S transition but also during S-phase and may even play a role in accelerating G2.
Unexpectedly, expression of CycD1 either during or after an aphidicolin block accelerates progression through S-phase into the next mitosis. We show directly that the associated CycD1 kinase complex is active during S and G2 phases (Figure 7). This notion is supported by the observation that Nicta;CycD3;3-associated kinases are active at both the G1/S and G2/M boundaries (Nakagami et al., 2002). Because B-type CDK genes are specifically expressed during S-phase and into M phase (Fobert et al., 1996), confirmed in our study for CDKB1 (Figure 4D), one cannot rule out that D-type cyclins could also interact B-type CDKs. It had been shown in vitro that Arath;CycD1;1 and Arath;CycD4;1, two D-type cyclins from Arabidopsis, can interact with CDKB1;1 and CDKB2;1 in addition to CDKA (Kono et al., 2003); however, in our study no interaction between ectopically expressed CycD1 and CDKB1 was observed in vivo.
Histone H1 and Rb proteins are known to be the substrates for plant D cyclin dependent kinases (Nakagami et al., 2002). A constitutively active nonphosphorylatable Rb protein can arrest rat cells in G1 and inhibit S-phase, indicating that Rb must be phosphorylated beyond G1/S for the completion of DNA replication (Knudsen et al., 1998). Cooper and Shayman (2001) also reported that phosphorylation of the Rb protein takes place throughout the cell cycle in actively cycling mammalian cells. Therefore, CDK activity may be required throughout S-phase to maintain and/or promote phosphorylation of Rb protein, ensuring a constant supply of active E2F transcription factor to promote the transcription of S-phase specific genes. Plant RBR-associated kinase complex contains both PSTAIRE CDKA and cyclin D2 (Boniotti and Gutierrez, 2001), but CDK complexes containing other cyclins are also capable of phosphorylating Rb (Nakagami et al., 2002). Indeed, we found that CycD1-associated kinase can phosphorylate both histone H1 and Rb proteins. The kinase activity toward Rb was found to increase continuously from an early S-phase until mitotic entry, suggesting that Rb phosphorylation is important not only for S but also for G2 phase progression. This findings are supported by the recent data from microarray analysis of transcription control by the E2F/Rb pathway in animal cells, which demonstrated more than 100 target genes, the majority of which are involved in cell cycle control, however, not only in DNA replication but also in G2/M transition and mitosis (Markey et al., 2002; Dimova et al., 2003).
CycD1 expression during G1 increased the total CDK activity judged by precipitated p13Suc1 beads. Increased kinase activity with distinct time courses for histone H1 and Rb proteins was caused by CycD1 expression also during the aphidicolin synchrony experiment. Rb kinase had smooth increase of activity toward its peak value preceding mitosis, and histone H1 kinase activity had an initial peak of activity followed by a decrease and second peak at the beginning of mitosis. The most likely mechanism of cell cycle promotion by CycD1 overexpression is through primary activation of the CDKA-CycD1 complex leading to a peak in its kinase activity toward histone H1 at 4 h after induction (Figures 5 and 7). Because the initial peak of total histone H1 kinase activity had been observed later, this probably represents a secondary enhancement of activity by phosphorylation cascade triggered by the CDKA-CycD1 complex. Therefore, D cyclins may be required continuously from G1 until mitotic entry. This is consistent with their continuous expression in proliferating cells (Sorrell et al., 1999), their interaction with both S-phase and mitotic CDKs (Nakagami et al., 2002), and their ability to reduce cell-doubling times (Cockcroft et al., 2000; Nakagami et al., 2002). However, the different groups of D cyclins appear to have different properties. When expressed in tobacco plants, Arath;CycD2;1 promoted cell cycle progression and growth by reducing G1 phase and promoting faster cell proliferation (Cockcroft et al., 2000). Overexpression of Arath;CycD3;1 in Arabidopsis had a distinctly different effect, prolonging the cell proliferation phase and inhibiting differentiation in developing leaves (Dewitte et al., 2003). Expression of Nicta;CycD3;3 in BY-2 cell culture reduced the proportion of cells in G1 phase as well as the cell-doubling times (Nakagami et al., 2002).
Do D Cyclins in Plants Have a More Diverse Range of Functions Than in Animals?
Animal D cyclins are thought to have a relatively restricted role, mediating entry into G1. Increasing the level of D or E cyclins leads to a shortening of the G1 phase in mammalian cells (Resnitzky and Reed, 1995). Inducible expression of D1 cyclin in breast cancer cells shortened G1 and was sufficient for cells arrested in G1 to enter and complete the cell cycle (Musgrove et al., 1994). These observations have been generally interpreted as indicating that these cyclins act during G1 phase, in part by activating cyclin E–dependent CDKs. Cyclin E is responsible for promoting S-phase entry and progression through S-phase. Cyclin D1 deficiency can be rescued by expression of cyclin E under the cyclin D promoter in mice (Geng et al., 1999), suggesting that cyclin E is a rate-limiting target for cyclin D1 in mammalian cells. D cyclins link the cell cycle to environmental conditions by responding to extracellular signals and acting via the Rb pathway leading to activation of cyclin E, which in turn regulates S-phase entry (Geng et al., 1999). There are no obvious cyclin E homologs in plants, but there is a much larger variety of D cyclins. The cyclin D family in Arabidopsis is large and seems to be functionally redundant—no mutant phenotype was observed for homozygous Arath;CycD3;2 knockout plants (Swaminathan et al., 2000). It is possible that plant D cyclins combine the functions performed by both D and E cyclins in animal cells and control G1 entry and progression into and through S-phase.
D1 cyclin differs from other D cyclins in many respects: it is usually found at much lower levels compared with D2 and D3 cyclins (Soni et al., 1995; Mironov et al., 1999), and sequence homology suggests it forms a distinct group from D2 or D3 (Renaudin et al., 1996). We have shown that expression of CycD1 accelerates cell cycle progression by promoting at least two distinct cell cycle phases. Our data suggests that, compared with mammalian D cyclins, CycD1 plays a fundamentally similar role as an accelerator of G1 to S-phase progression. However, plant CycD1 has a substantially different role during S, and possibly G2, phases. CycD1 accelerates G1/S and S/G2, whereas overexpression of animal D cyclin promotes only transition from G1 to S-phase (Sherr and Roberts, 1999; Shen, 2002). This dual role could be a common feature of plant D cyclins because ectopic expression of Arath;CycD3;1 induced not only DNA replication but also cell division in trichomes (Schnittger et al., 2002). The interpretation of D cyclin function in maintenance of fast proliferation is consistent with our data on growth rate in induced cultures.
Stacey (2003) proposed a model, based on studies of actively cycling mouse and human cell cultures using time-lapse photography and quantitative image analysis, whereby mammalian D1 cyclin is required for the transition from G1 to S-phase and initiation of DNA synthesis. However, to complete DNA synthesis, D1 cyclin must be degraded. A decline in D1 cyclin levels during S-phase (Pagano et al., 1994) is necessary for proliferating cell nuclear antigen relocation and DNA repair/synthesis as well as for the start of DNA replication. After the completion of S-phase and only if conditions are conducive for continued growth, D1 cyclin levels can increase during G2 phase. The accumulation of D1 cyclin during G2 determines if the cell will continue through the next cell cycle. However, when conditions are not conducive for continued proliferation, D1 cyclin levels remain low during G2 phase without any effect on timing of entry into the current mitosis. Thus, D1 cyclin behaves as a switch that has to be turned off in S-phase but can be turned on again in G2 phase for continued proliferation (Stacey, 2003). According to this model, the levels of D1 cyclin should be lowest during S-phase.
Our data indicate that, in plants, CycD1 degradation is not required for S-phase progression. Indeed, CycD1 is stable up until mitosis, its associated kinase is active during G2 but inactivated after mitosis (Figure 5D), and ectopic expression during S-phase actually enhances cell cycle progression. Therefore, we conclude that plant and mammalian D cyclins can both facilitate the G1/S transition, but their role in further cell cycle progression have diverged significantly.
METHODS
Transformation of Cell Cultures
BY-2 (Bright Yellow) tobacco (Nicotiana tabacum) cells, grown in the dark at 25°C and shaken at 150 rpm, were subcultured weekly by transferring 2-mL aliquots to 100 mL of fresh media (Nagata et al., 1992). Using inducible swith AlcR:GR, which is the translational fusion of AlcR transcription factor to GR transactivator domain (G. Roberts, O. Koroleva, M. Ito, P. Laufs, D. Leader, and J. Doonan, unpublished data), allowed us to use induction of gene expression under AlcA promoter by Dex instead of ethanol used in the original Alc system (Roslan et al., 2001). Transgenic lines of BY-2 carrying the receptor switch cassette AlcR:GR, were retransformed with a second cassette containing the CycD1 gene or tagged HA:CycD1 under control of the AlcA promoter (Parinyapong, 2002), using a hypervirulent strain of Agrobacterium tumefaciens (LBA4404.pBBR1MCSvirGN54D; van der Fits et al., 2000) and a transformation protocol described by An (1985).
For protein localization studies, GFP was translationally fused to the N terminus of CycD1 using the GATEWAY system (Invitrogen, Carlsbad, CA) and pGWB6 expression vector (gift of T. Nakagawa, Shimane University, Japan), and both were transiently expressed under the CaMV35S promoter in Arabidopsis thaliana suspension culture (Mathur et al., 1998) and transformed for stable expression into BY-2 cells using hypervirulent strain of Agrobacterium (as described above).
Cell Synchronization and Induction
Release from stationary phase into G1/S was achieved by diluting a 10-d-old culture with fresh media in a 1:10 ratio. To monitor the effect of inducible CycD1 expression, each diluted culture was split equally into two sterile flasks, and water-soluble Dex was added to a final concentration of 10 μM to one of the flasks. The other was used as a noninduced control.
Aphidicolin synchrony was performed essentially as described by Nagata et al. (1992). A 7-d-old culture was diluted with fresh media in a ratio of 1:7, and aphidicolin (Sigma, St. Louis, MO) was added to each at a concentration of 5 μg/mL.
Flow Cytometry Analysis and Determination of MI
Samples (5 mL) for flow cytometry were centrifuged for 3 min at 700 rpm, and the pellets were immediately frozen in liquid nitrogen and kept at −80°C before analysis. The frozen pellets (∼100 to 200 mg) were thawed with gentle agitation in 300 μL of Partec extraction buffer, and after adding 1.5 mL of Partec staining buffer, the suspension was filtered through 30-μM nylon mesh filters and analyzed on a Partec PAS II flow cytometer (Partec, Munster, Germany). Cell cycle data were analyzed using FloMax software (Partec) and Multicycle for Windows (Phoenix Flow Systems, San Diego, CA). Both methods of analysis produced similar results.
The MI was determined by fixing samples in 4% (w/v) paraformaldehyde, 10 mM EDTA, 5 mM MgSO4, and 0.1 M sorbitol in a 50 mM Pipes buffer, pH 7.0, staining with 2 μg/mL 4′,6-diamidino-2-phenylindole in the presence of 1% (v/v) Igepal detergent (Sigma), and observing under an epifluorescence microscope. At least 800 cells for each time point were scored for mitotic figures when a single culture of each treatment was analyzed. When experiments were run in triplicate for each treatment, for each time point at least 400 cells from each flask (=1200 per treatment) were analyzed.
mRNA Isolation and Analysis by PCR
Oligo(dT) Dynabeads (Dynal, Bromborough, UK) were used to isolate mRNA. Approximately 50 mg of cell pellet, frozen in liquid nitrogen, was ground in 100 μL of lysis/binding buffer (Dynal) and centrifuged for 1 min at 13,000 rpm in a benchtop centrifuge. Fifty microliter of supernatant was added to 20 μL of prepared Dynabeads and incubated and washed according to manufacturer's instructions. Reverse transcription was performed using the reaction mixture Omniscript (Qiagen, Valencia, CA). PCR reactions were performed in a reaction volume of 25 μL (Taq Master Mix; Qiagen) with 4 μL of the cDNA reaction mixture containing suspended magnetic beads. NtGAPDH amplification was used as loading control. The oligonucleotide primer pairs had the following sequences: CycD1-forward 5′-CTAAACGCCCATCACTTGCCG-3′ and CycD1-reverse 5′-TTGTCATCATCCGCCCGAGAGC-3′ (amplifying a fragment of 653 bp); NtGAPDH-forward 5′-GGTTTGGCATTGTGGAGGGTC-3′ and NtGAPDH-reverse 5′-CCCTCCGATTCCTCCTTGATTGC-3′ (amplifying a fragment of 304 bp); NtCycD2;1 and NtCycD3;2 as described by Sorrell et al. (1999). After 25 PCR cycles with primer annealing temperature of 55°C, 10-μL samples of the PCR reaction mixture were separated on a 1% (w/v) agarose gel (Figure 2B).
Protein Gel Blot Analysis
Cultures in midexponential phase of growth (4 d after dilution) were induced by adding Dex to a final concentration of 10 μM. After 0.5, 1, 2, or 24 h, 1.5-mL samples were centrifuged at 700 rpm for 3 min, and the cell pellet was frozen in liquid nitrogen. Protein was extracted by grinding in an equal volume of sample buffer (62.5 mM Tris-HCl, pH 6.8, containing 5% [v/v] β-mercaptoethanol, 2% [w/v] SDS, 10% [v/v] glycerol, and 0.001% [w/v] bromophenol blue). Immunoblotting was performed essentially as described by Towbin and Gordon (1984), and specific proteins were detected using monoclonal mouse anti-HA antibody (used at 1:1,000 dilution; HA.11; Covance, Berkeley, CA), anti-PSTAIR (used at 1:5,000 dilution; Sigma), and secondary anti-mouse horseradish peroxidase–conjugated antibody (used at 1:10,000 dilution; DAKO, Ely, UK) and detected with Super Signal West Pico chemiluminescent substrate (Pierce, Rockford, IL). Equivalent protein loading for selected samples was achieved by normalization of loading after measurement of protein content by Bradford reagent (Sigma) and verified either by Ponceau S (Sigma) staining before immunostaining or by immunoblotting of native proteins (i.e., CDKA and tubulin).
Immunoprecipitation
Cultures in the exponential phase of growth (4 d after dilution by fresh media) were induced with 10 μM Dex for 24 h as described above. The proteasome inhibitor MG132 was added to some cultures at a final concentration of 100 μM 2 h before protein extraction. For each treatment, 50 mL of culture was centrifuged 3 min at 1000 rpm at 4°C, the cell pellet frozen in liquid nitrogen, ground and mixed with an equal volume of extraction buffer (50 mM Tris-HCl, 150 mM NaCl, 15 mM EGTA, 15 mM MgCl2, 1 mM DTT, and 0.1% [v/v] Tween 20, pH 7.5) containing a protease-inhibitor cocktail (Roche, Indianapolis, IN; one tablet/10 mL), then centrifuged 20 min at 38,000g at 4°C. For immunoprecipitation, 1 mL of each protein extract (∼4 mg of total protein) was added to 100 μL of beads (anti-HA agarose [Roche] or anti-goat IgG agarose [Sigma]) and incubated at 4°C for 16 h with gentle agitation and then washed four times with the extraction buffer and finally resuspended in 150 μL of sample buffer, heated 5 min at 80°C, separated on SDS-PAGE, and immunoblotted as described above.
Kinase Assays
Samples (1.5 mL) were taken at discrete time points from cultures in G1 or aphidicolin synchrony experiments and centrifuged at 3000 rpm for 3 min at 4°C. Cell pellets were frozen in liquid nitrogen, then ground in 500 μL of the kinase buffer (Nakagami et al., 2002) and cleared by centrifugation (14,000 rpm) for 20 min at 4°C. Agarose beads conjugated with either anti-HA monoclonal antibody (Roche) or p13Suc1 protein (gift from L. Bogre) were washed once and then resuspended in the same volume of kinase buffer or Suc1 buffer (50 mM Tris-HCL, pH 7.5, 250 mM NaCl, 5 mM EGTA, 5 mM EDTA, 0.1% Tween 20, 5 mM NaF, 0.1% Igepal CA 630, and 0.5 mM phenylmethylsulfonyl fluoride), respectively. Five hundred microliters of cell extract were added to 20 μL of beads in Eppendorf tubes and incubated 2 h at 4°C with gentle rotation. The beads were then collected by a short spin and washed by kinase buffer (HA beads) or Suc1 buffer (Suc1 beads) 4 × 500 μL, with final wash by kinase buffer for both types of beads. Kinase assays were started by adding 10 μL of kinase buffer containing 100 μM ATP, 74 kBq of γ-32P-ATP (Amersham, Buckinghamshire, UK) and either 1 μg/μL of histone H1 (Roche) or GST-NtRb1 fusion protein preparation from Escherichia coli expressing GST-NtRb1 fragment in pGEX-4T-1 (gift from M. Sekine). The reaction mixtures were incubated at room temperature for 30 min and terminated by the addition of 5 μL of 4× SDS sample buffer heated at 95°C for 5 min and analyzed by SDS-PAGE with subsequent autoradiography. Quantitation of band integrated optical density was performed using LabWorks image acquisition and analysis software (Ultra-Violet Products, Cambridge, UK).
Growth Analysis Using Bioscreen C
A Bioscreen C plate reader shaker incubator (Thermo Electron Corporation, Waltham, MA) supplied with Growth Curves software (Transgalactic, Helsinki, Finland) was used to monitor optical density (OD600) of cell suspensions. Stationary cultures were diluted 1:20 with fresh media and split onto two flasks, and Dex was added to one of the flasks at a final concentration of 5 μM. The cell suspensions were thoroughly mixed and aliquoted 200 μL per single well of a honeycomb plate, with 10 (wells) replicas for each treatment. Plates were inserted into the incubation chamber of the Bioscreen C and run at 25°C, with continuous extra-intensive shaking and OD600 measurements performed every hour.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AJ250396.
Acknowledgments
We are grateful to A.P. Hutchins and G.R. Roberts for valuable advice and discussions; to Margit Menges for advice on flow cytometric analysis; to Lazlo Bogre for the gift of p13Suc1 agarose beads; to Masami Sekine for the gift of NtCDKB1antibody and GST-NtRb1 plasmid. This work was funded by a Syngenta grant (O.A.K., M.T., and L.S.), a grant-in-aid from Biotechnology and Biological Science Research Council to the John Innes Centre, and by a PhD studentship (Ministry of Science, Technology, and Environment and the Royal Thai Government) to P.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John H. Doonan (john.doonan@bbsrc.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023754.
References
- An, G. (1985). High efficiency transformation of cultured tobacco cells. Plant Physiol. 79, 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ach, R.A., Durfee, T., Miller, A.B., Taranto, P., Hanley-Bowdoin, L., Zambryski, P.C., and Gruissem, W. (1997). RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Augustin, J.C., Rosso, L., and Carlier, V. (1999). Estimation of temperature dependent growth rate and lag time of Listeria monocytogenes by optical density measurements. J. Microbiol. Methods 38, 137–146. [DOI] [PubMed] [Google Scholar]
- Boniotti, M.B., and Gutierrez, C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28, 341–350. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Cooper, S. (1998). On the interpretation of the shortening of the G1-phase by overexpression of cyclins in mammalian cells. Exp. Cell Res. 238, 110–115. [DOI] [PubMed] [Google Scholar]
- Cooper, S., and Shayman, J.A. (2001). Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell. Mol. Life Sci. 58, 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova, D.K., Stevaux, O., Frolov, M.V., and Dyson, N.J. (2003). Cell cycle-dependent and cell cycle-independent control of transcription by Drosophila E2F/RB pathway. Genes Dev. 17, 2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam, V.M., Orrechia, S., Lowe, M., Pardi, R., and Pines, J. (2001). The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 152, 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert, P.R., Gaudin, V., Lunness, P., Coen, E.S., and Doonan, J.H. (1996). Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, D., Riou-Khamlichi, C., Oakenfull, E.A., and Murray, J.A. (2003). Isolation, characterization and expression of cyclin and cyclin-dependent kinase genes in Jerusalem artichoke (Helianthus tuberosus L.). J. Exp. Bot. 54, 303–308. [DOI] [PubMed] [Google Scholar]
- Fuerst, R.A., Soni, R., Murray, J.A., and Lindsey, K. (1996). Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 112, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin, V., Lunness, P.A., Fobert, P.R., Towers, M., Riou-Khamlichi, C., Murray, J.A.H., Coen, E., and Doonan, J.H. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the cycloidea gene. Plant Physiol. 122, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, Y., Whoriskey, W., Park, M.Y., Bronson, R.T., Medema, R.H., Li, T., Weinberg, R.A., and Sicinski, P. (1999). Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97, 767–777. [DOI] [PubMed] [Google Scholar]
- Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J.E., Bhattacharya, S., Rideout, W.M., Bronson, R.T., Gardner, H., and Sicinski, P. (2003). Cyclin E ablation in the mouse. Cell 114, 431–443. [DOI] [PubMed] [Google Scholar]
- Healy, J.M.S., Menges, M., Doonan, J.H., and Murray, J.A.H. (2001). The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J. Biol. Chem. 276, 7041–7047. [DOI] [PubMed] [Google Scholar]
- Huntley, R., et al. (1998). The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol. 37, 155–169. [DOI] [PubMed] [Google Scholar]
- Huntley, R.P., and Murray, J.A.H. (1999). The plant cell cycle. Curr. Opin. Plant Biol. 2, 440–446. [DOI] [PubMed] [Google Scholar]
- Jackman, M., Lindon, C., Nigg, E.A., and Pines, J. (2003). Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5, 143–148. [DOI] [PubMed] [Google Scholar]
- Knudsen, E.S., Buckmaster, C., Chen, T.T., Feramisco, J.R., and Wang, J.Y. (1998). Inhibition of DNA synthesis by RB: Effects on G1/S transition and S-phase progression. Genes Dev. 12, 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, A., Umeda-Hara, C., Lee, J., Ito, M., Uchimiya, H., and Umeda, M. (2003). Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132, 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey, M.P., Angus, S.P., Strobeck, M.W., Williams, S.L., Gunawardena, R.W., Aronow, B.J., and Knudsen, E.S. (2002). Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 62, 6587–6597. [PubMed] [Google Scholar]
- Mathur, J., Szabados, L., Schaefer, S., Grunenberg, B., Lossow, A., Jonas-Straube, E., Schell, J., Koncz, C., and Koncz-Kalman, Z. (1998). Gene identification with sequenced T-DNA tags generated by transformation of Arabidopsis cell suspension. Plant J. 13, 707–716. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. [DOI] [PubMed] [Google Scholar]
- Minshull, J., Blow, J.J., and Hunt, T. (1989). Translation of cyclin mRNA is necessary for extracts of activated xenopus eggs to enter mitosis. Cell 56, 947–956. [DOI] [PubMed] [Google Scholar]
- Mironov, V.V., De Veylder, L., Van Montagu, M., and Inze, D. (1999). Cyclin-dependent kinases and cell division in plants—The nexus. Plant Cell 11, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motokura, T., and Arnold, A. (1993). Cyclin D and oncogenesis. Curr. Opin. Genet. Dev. 3, 5–10. [DOI] [PubMed] [Google Scholar]
- Musgrove, E.A., Lee, C.S., Buckley, M.F., and Sutherland, R.L. (1994). Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc. Natl. Acad. Sci. USA 91, 8022–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY-2 cell-line as the HeLa-cell in the cell biology of higher plants. Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- Nakagami, H., Kawamura, K., Sugisaka, K., Sekine, M., and Shinmyo, A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami, H., Sekine, M., Murakami, H., and Shinmyo, A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 18, 243–252. [DOI] [PubMed] [Google Scholar]
- Oakenfull, E.A., Riou-Khamlichi, C., and Murray, J.A. (2002). Plant D-type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, M., Theodoras, A.M., Tam, S.W., and Draetta, G.F. (1994). Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 8, 1627–1639. [DOI] [PubMed] [Google Scholar]
- Parinyapong, P. (2002). An Analysis of Antirrhinum majus Cyclin A20 and Cyclin D1 Function in Arabidopsis thaliana. PhD dissertation (Liverpool, UK: University of Liverpool).
- Pines, J. (1995). Cyclins and cyclin-dependent kinases—A biochemical view. Biochem. J. 308, 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais, S., Glab, N., Trehin, C., Perennes, C., Bureau, J.M., Meijer, L., and Bergounioux, C. (1997). Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 12, 191–202. [DOI] [PubMed] [Google Scholar]
- Renaudin, J.P., et al. (1996). Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol. 32, 1003–1018. [DOI] [PubMed] [Google Scholar]
- Resnitzky, D., and Reed, S.I. (1995). Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol. 15, 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20, 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S., Wells, R., and Rechsteiner, M. (1986). Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 234, 364–368. [DOI] [PubMed] [Google Scholar]
- Roslan, H.A., Salter, M.G., Wood, C.D., White, M.R., Croft, K.P., Robson, F., Coupland, G., Doonan, J., Laufs, P., Tomsett, A.B., and Caddick, M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28, 225–235. [DOI] [PubMed] [Google Scholar]
- Sala, F., Galli, M.G., Nielsen, E., Magnien, E., Devreux, M., Pedrali-Noy, G., and Spadari, S. (1983). Synchronization of nuclear DNA synthesis in cultured Daucus carota L. cells by aphidicolin. FEBS Lett. 153, 204–208. [Google Scholar]
- Sewing, A., Burger, C., Brusselbach, S., Schalk, C., Lucibello, F.C., and Muller, R. (1993). Human cyclin D1 encodes a labile nuclear protein whose synthesis is directly induced by growth factors and suppressed by cyclic AMP. J. Cell Sci. 104, 545–555. [DOI] [PubMed] [Google Scholar]
- Shen, W.H. (2002). The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 7, 505–511. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J. (1994). G1 phase progression: Cycling on cue. Cell 79, 551–555. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1999). CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.D., and Hulskamp, M. (2002). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99, 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., Shah, Z.H., and Murray, J.A. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell, D.A., Combettes, B., Chaubet-Gigot, N., Gigot, C., and Murray, J.A. (1999). Distinct cyclin D genes show mitotic accumulation or constant levels of transcripts in tobacco bright yellow-2 cells. Plant Physiol. 119, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell, D.A., Menges, M., Healy, J.M., Deveaux, Y., Amano, C., Su, Y., Nakagami, H., Shinmyo, A., Doonan, J.H., Sekine, M., and Murray, J.A. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol. 126, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, D.W. (2003). Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 15, 158–163. [DOI] [PubMed] [Google Scholar]
- Swaminathan, K., Yang, Y., Grotz, N., Campisi, L., and Jack, T. (2000). An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol. 124, 1658–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., and Gordon, J. (1984). Immunoblotting and dot immunobinding—Current status and outlook. J. Immunol. Methods 72, 313–340. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits, L., Deakin, E.A., Hoge, J.H., and Memelink, J. (2000). The ternary transformation system: Constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 43, 495–502. [DOI] [PubMed] [Google Scholar]
- Weingartner, M., Binarova, P., Drykova, D., Schweighofer, A., David, J.P., Heberle-Bors, E., Doonan, J., and Bogre, L. (2001). Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13, 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]