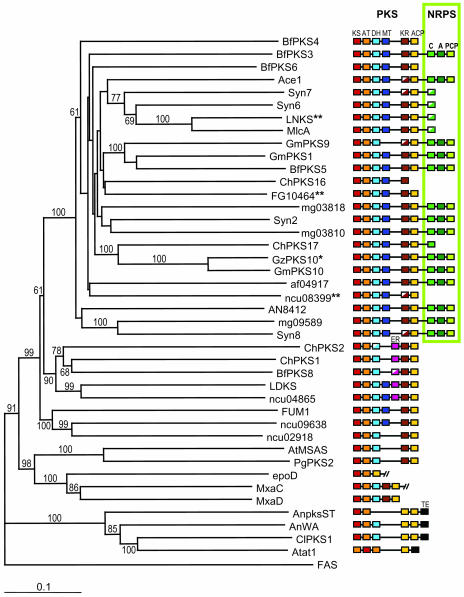
Figure 3.
Phylogeny of Fungal PKS Based on Protein Sequence of KS and AT Domains.
The consensus phylogenetic tree was obtained using the neighbor-joining method with distances. Bootstrap values are indicated above or below nodes of the tree. Colored boxes represent PKS and NRPS enzymatic domains (see Figure 2 for definitions). Sequences from the following organisms were included in this analysis (see Methods for accession numbers): M. grisea (Ace1, Syn2, Syn6,7,8, mg03810, mg03818, and mg09589); A. fumigatus (af04917); A. nidulans (AN8412, AnpksST, and AnWA); F. graminearum (FG10464); N. crassa (ncu08399, ncu04865, ncu09638, and ncu02918); A. terreus (LNKS, LDKS, AtMSAS, and Atat1); P. citrinum (MlcA); P. griseofulvum (PgPKS2), C. heterostrophus (ChPKS1); G. moniliformis (FUM1), and C. lagenarium (ClPKS1). We added fungal PKS and PKS/NRPS sequences described by Kroken et al. (2003) from C. heterostrophus (ChPKS2,16,17), B. fuckeliana (BfPKS3,4,5,6,8), G. moniliformis (GmPKS1,9,10), and G. zeae (GzPKS10) and bacterial PKS sequences (epoD, MxaC, and MxaD) as references. FAS from the silk moth Bombyx mori was included as an outgroup to root the tree and PKS/NRPS cluster as a monophyletic group. *, Although GzPKS10 is only described as a partial PKS sequence, its relatedness to GmPKS10 (90% identity) suggests that it has a full NRPS module as GmPKS10. **, FG10464 is almost identical (>91% identity) to GzPKS9 from Kroken et al. (2003). ncu08399 is identical to NcPKS4 from Kroken et al. (2003). LNKS is identical to lovB from Kroken et al. (2003).