Figure 5.
Silencing by a Posttranscriptional Mechanism.
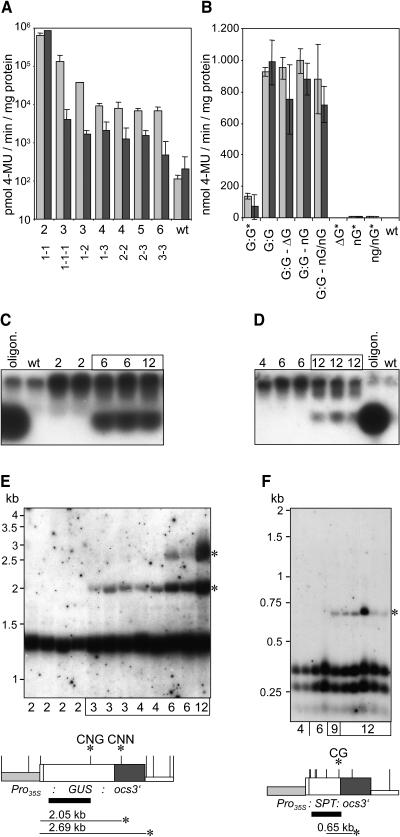
(A) GUS activity assays revealed silencing of the GUS gene in transgenic lines harboring three or more copies of the GUS reporter gene under the control of the CaMV 35S promoter. Extracts were prepared from F1 progeny plants derived from crosses among homozygous transgenic lines harboring T-DNAs GUS 1xr, GUS 2xr, and GUS 3xr. The results for 2-week-old and 8-week-old plants are shown as light and dark gray bars, respectively. Copy numbers of GUS genes in the genome and at the different T-DNA loci are indicated for the different plants. Dashes divide copies of different loci.
(B) Lowly expressed GUS genes do not trigger transgene silencing. Either transgenic lines homozygous for a particular T-DNA were analyzed, or F1 progeny plants derived from crosses of homozygous transgenic lines among each other or with wild-type plants. Light and dark gray bars indicate the results for 2-week-old and 8-week-old plants. T-DNA loci GUS 2*xr 114, ΔGUS 1xr 102, NOS-GUS 1xr 111, and NOS-GUS 2xr 108 are abbreviated G:G, ΔG, nG, and nG/nG, respectively. Construct GUS 2*xr is a derivative of construct GUS 2xr that contains the SPT reporter gene cassette as a spacer between two GUS reporter gene cassettes under the control of the CaMV 35S promoter in a tandem arrangement. Different loci are divided by dashes, and asterisks mark plants homozygous for a particular T-DNA.
(C) and (D) Detection of siRNAs in 8-week-old plants of lines displaying silencing of the GUS (C) or SPT gene (D). A GUS-oligonucleotide (20 nucleotides, 75 ng) and an ocs-oligonucleotide (21 nucleotides, 75 ng) mixed with wild-type RNA served as size standard and hybridization control for the detection of siRNAs with probes corresponding to the GUS reporter gene and ocs3′ sequences, respectively. Numbers of transgene copies in the genome are given for each line.
(E) and (F) Methylation analysis of plants carrying different copy numbers of the GUS (E) or SPT genes (F). DNA of 8-week-old plants harboring GUS and SPT genes was cut with AluI and Bsp143II, respectively. The resulting DNA gel blots were hybridized with the probes that are indicated as solid black bars. AluI and Bsp143II restriction sites are shown as vertical bars on the maps of the GUS and the SPT reporter gene cassettes, respectively. Asterisks indicate hybridizing fragments indicative of methylation as well as the methylated sites. It is shown whether CG, CNG, or CNN methylation took place.