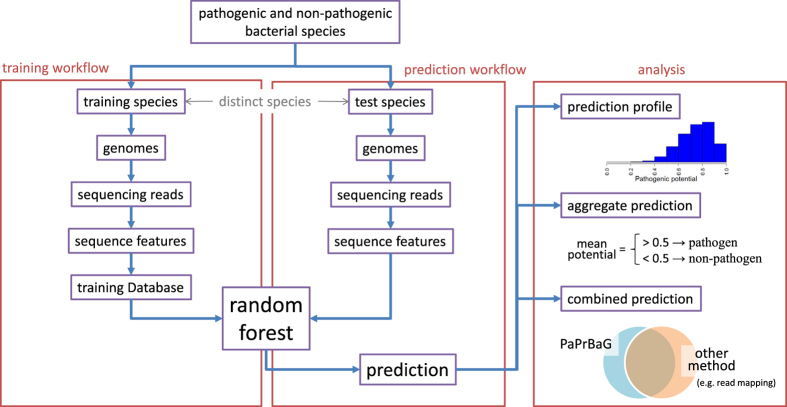
Figure 1. Overview of PaPrBaG workflow.
Reads are simulated from genomes in both the training (left) and prediction (center) workflows, from which features are extracted. The training sequence features together with the associated phenotype labels compose the training database, on which the random forest algorithm trains a pathogenicity classifier. This classifier predicts the pathogenic potential for each read in the test set. From these raw results, the prediction profile, the genome aggregate prediction and a combined prediction can be generated (right).