Fig. 1.
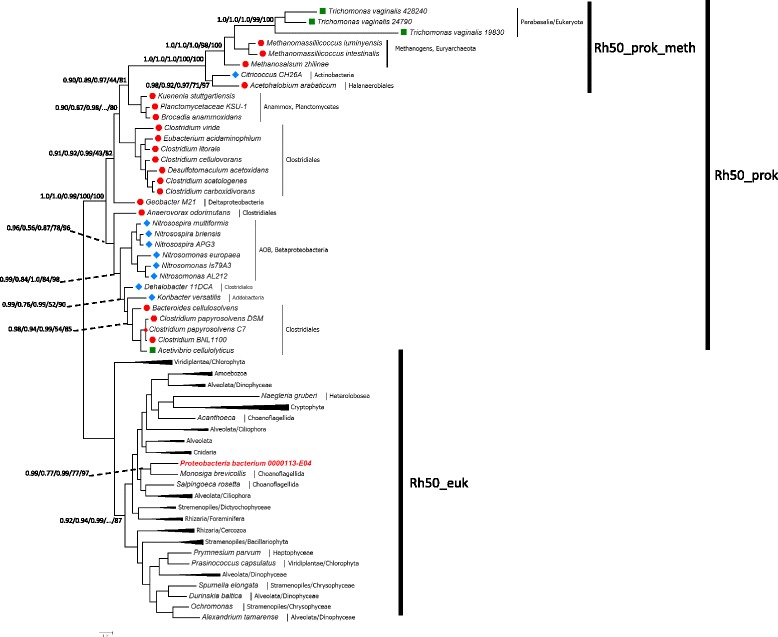
Unrooted phylogeny of Rh50 proteins (90 taxa, 252 sites). Bayesian majority-rule consensus of 1,076 trees obtained under the LG + Γ4 mixture model in PhyloBayes. Branch-support values given at nodes are PP1/PP2/aBayes/RBS/UFBoot. PP1 = Bayesian Posterior Probabilities (LG + Γ4; PhyloBayes), PP2 = Bayesian Posterior Probabilities (CATGTR + Γ4; PhyloBayes); aBayes = Maximum likelihood Bayesian-like aLTR transformation (LG + Γ4 + F; PhyML); RBS = Maximum likelihood rapid-bootstrap support (LG + Γ4 + F; RAxML); UFBoot = Maximum likelihood ultrafast-bootstrap approximation (LG + Γ4 + F; IQ-TREE). Some clades are condensed for clarity of presentation (for full tree see Additional file 2: Figure S1). Species oxygen requirements are denoted as follows: anaerobic (red dot), facultative (green square), aerobic (blue diamond). Proteobacteria bacterium JGI 0000113-E04 is highlighted in red. The genome of Proteobacteria bacterium JGI 0000113-E04 is deposited at the IMG-JGI (GOLD project ID = Gp0024568). Several lines of evidence suggest that part of the scaffolds making up this genome belong to contaminants. The RH50 homolog (Locus tag = D341DRAFT_01802) is present at one end of scaffold00071.71. Its nearest neighbour on the scaffold (D341DRAFT_01804), which codes for a beta-galactosidase/beta-glucuronidase, is again sister to its Monosiga homolog in a Bayesian phylogeny (Additional file 2: Figure S2). The P. bacterium genome is issued from uncultured single-cell sequencing, and no genomic DNA is available, thus experimental tests cannot be performed. The scale-bar indicates the estimated number of substitutions per site
