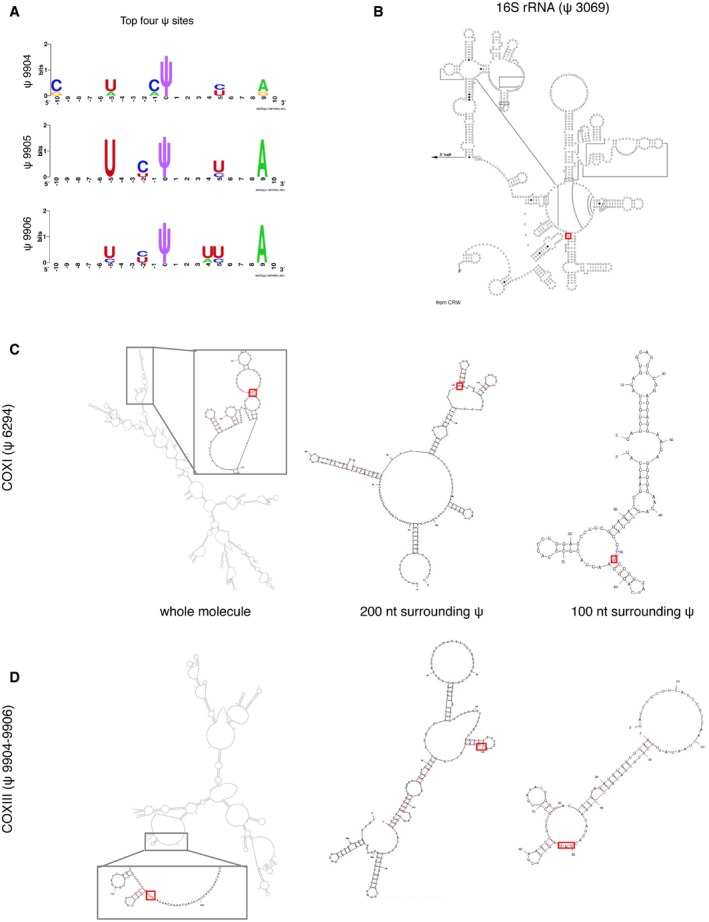
Positions of identified ψ sites (red rectangle) within the secondary structures of individual RNAs. (B) Known secondary structure of the 3′‐terminus of 16S rRNA
37 and predicted structures for COXI (C) and COXIII mRNAs (D). Predictions were based either on sequence fragments of 100 nt and 200 nt surrounding each ψ site, or using the whole mRNA molecule as the input. Pseudouridine is predicted to be accessible in all cases, located at a loop or in the boundary between a stem and a loop/bulge structure.