Figure EV3. Chromatin compaction or kinetochore closeness to SPBs is not affected in cells lacking Cdc14 activity in response to a DSB.
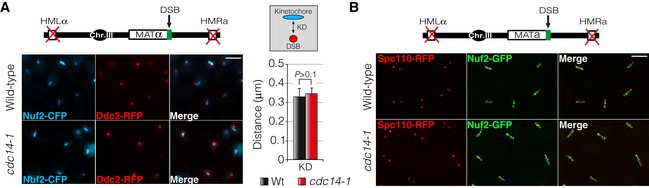
- Chromatin compaction during the DDR is not affected in the absence of Cdc14. A single DSB was generated by expressing the HO endonuclease in both wild‐type and cdc14‐1 mutant cells. Nuf2‐CFP and Ddc2‐RFP were used as kinetochore and DSB markers, respectively. Micrographs were generated by using the maximum projection of nine z‐planes images. Scale bar: 3 μm. The kinetochore–DSB distance in both wild‐type and cdc14‐1 backgrounds was measured and plotted. Graph depicts the mean ± SD of three replicates of at least 100 cells each. P‐value was calculated using a two‐tailed unpaired Student's t‐test.
- Cdc14 is not required to promote DSB‐SPB interaction by generating pulling forces that move kinetochores regions close to the SPBs during the DNA damage response. A single DSB was produced by inducing the HO endonuclease expression in both wild‐type and cdc14‐1 mutant cells. Spc110‐RFP and Nuf2‐GFP were used to visualize the SPBs and the kinetochore, respectively. Micrographs were created by using the maximum projection of nine z‐planes images. Scale bar: 3 μm.
