Figure 1.
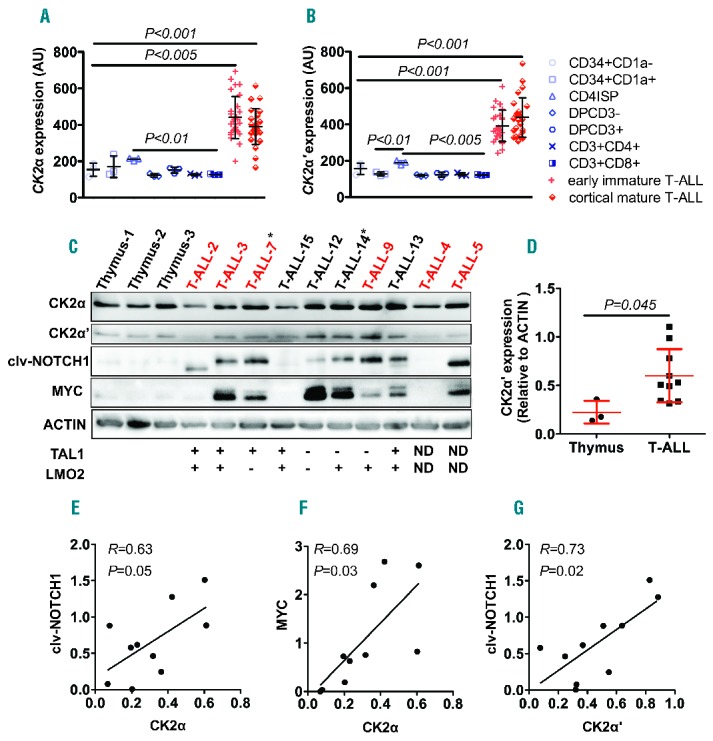
CK2 expression is elevated in human patient T-cell acute lymphoblastic leukemia (T-ALL) cells, and correlates with those of NOTCH1 and MYC. (A and B). Both CK2α (probe ID: ILMN_2386355) (A) and CK2α′ (probe ID: ILMN_1723843) (B) transcripts are elevated in early immature and mature T-ALL patient samples, compared with different subsets of T cells. Mean±SD of CK2α: 440.6±21.9 for early immature T-ALL and 391.1±19.6 for cortical/mature T-ALL versus 154±21.1 for CD34+CD1a−, 170.1±34.35 for CD34+CD1a+, 211.1±4.55 for CD4ISP, 122.3±5.49 for DPCD3−, 151.5±9.28 for DPCD3+, 126.3±3.19 for CD3+CD4+, 127.1±2 for CD3+CD8+; P<0.005 and P<0.001; and CK2α′: 392.2±16.3 for early immature T-ALL and 438.1±21.74 for cortical/mature T-ALL versus 156.8±17.89 for CD34+CD1a−, 128.1±5.5 for CD34+CD1a+, 190±6.81 for CD4ISP, 119.4±2.61 for DPCD3−, 124.1±7.54 for DPCD3+, 124.6±5.18 for CD3+CD4+, 122.4±1.94 for CD3+CD8+. P<0.001 for all comparisons; n=28 for early immature T-ALL, 25 for cortical/mature T-ALL and 3 for subsets of T cells, respectively. Among the different T-cell subsets, the expression of CK2α (A) and CK2α′ (B) is slightly but significantly higher in CD4ISP cells, compared to double-positive or single-positive subsets of T cells (P<0.01 and P<0.005, respectively). (C) Western blotting analysis of CK2α, CK2α′, cleaved-NOTCH1 (clv-NOTCH1) and MYC in patient T-ALL samples, compared with normal thymus. ACTIN serves as a loading control. Patient sample number marked in red indicates NOTCH1 mutations and asterisk (*) denotes FBW7 mutations. (D) CK2α′ versus ACTIN protein ratios demonstrating that CK2α′ levels are significantly higher in primary T-ALL patient samples, compared with those in control thymocytes (mean±SD of CK2α′ to ACTIN ratio: 0.60±0.09 versus 0.22± 0.07; P=0.045; n=10 and 3, respectively). (E–G) Pearson correlation tests reveal that CK2α (E and F) and CK2α′ (G) protein levels significantly correlate with those of NOTCH1 and MYC, or NOTCH1 alone (n=10; P=0.05, 0.03 and 0.02, respectively). AU: arbitrary unit. All human samples were collected and analyzed after informed consent and with approval of the Institutional Review Board and the Ethics Committee without linked identifiers.
