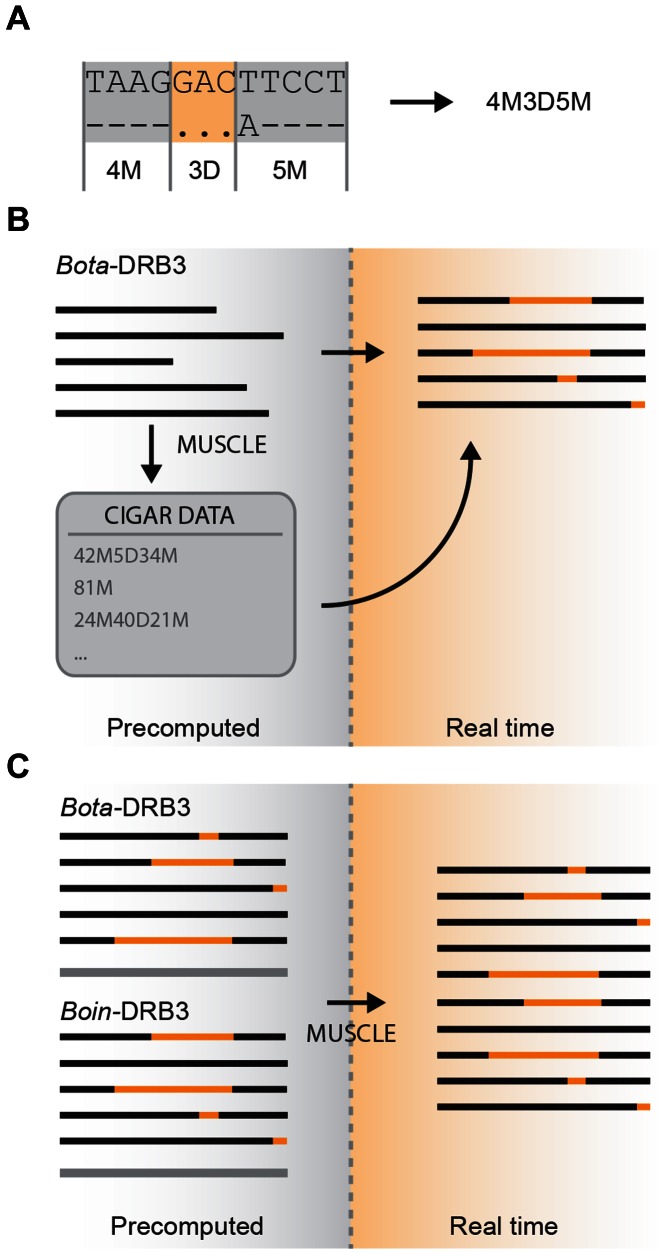
Figure 2.
Single- and multi-locus alignment. (A) For each computed alignment, a CIGAR (Compact Idiosyncratic Gapped Alignment Report) string defining the sequence of matches/mismatches (M) and deletions or gaps (D) compared to the reference sequence is stored in the database. (B) For each locus in the database, the nucleotide and protein allele alignment is pre-computed and the CIGAR string is stored in the database to correctly represent the sequence alignment. (C) In multi-locus alignment, the consensus sequence of each locus is aligned in real time and the previously calculated single-locus aligned are assembled and rendered as one.