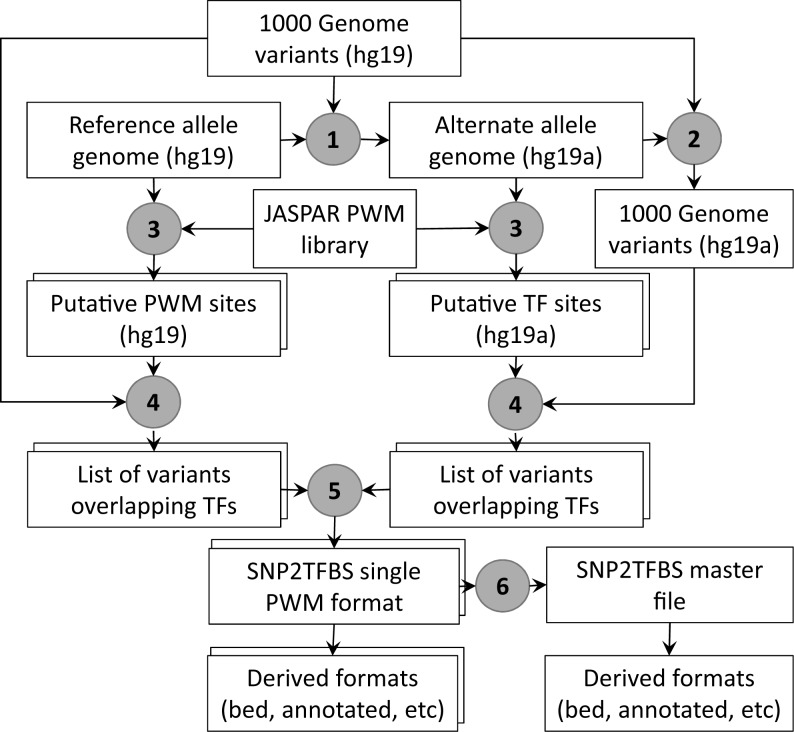
Figure 1.
SNP2TFBS data generation pipeline. The rectangular boxes represent data files. Duplicated frames indicate multiple files for each type, one for each position weight matrice (PWM). Encircled numbers refer to procedures: (1) Generation of the alternate human genome (hg19a) from reference genome (hg19) (2) Genome coordinates conversion of the reference single nucleotide polymorphisms (SNP) catalogue (VCF format) to the alternate genome. (3) Whole genome scan of both genomes with PWMs from JASPAR CORE 2014 at P-value threshold 10−5.(4) Extraction of SNPs overlapping PWM matches on the respective genomes. (5) Extraction of SNPs that disrupt, create or change score of overlapping PWM sites between the two genomes (6) Merging of essential information from single PWM files into master file, generation of gene-annotated and reformatted versions (e.g. BED) from primary data files. Variants annotation is carried out using an ANNOVAR input file (humandb/hg19_refGene.txt).