Abstract
The human disease methylation database (DiseaseMeth, http://bioinfo.hrbmu.edu.cn/diseasemeth/) is an interactive database that aims to present the most complete collection and annotation of aberrant DNA methylation in human diseases, especially various cancers. Recently, the high-throughput microarray and sequencing technologies have promoted the production of methylome data that contain comprehensive knowledge of human diseases. In this DiseaseMeth update, we have increased the number of samples from 3610 to 32 701, the number of diseases from 72 to 88 and the disease–gene associations from 216 201 to 679 602. DiseaseMeth version 2.0 provides an expanded comprehensive list of disease–gene associations based on manual curation from experimental studies and computational identification from high-throughput methylome data. Besides the data expansion, we also updated the search engine and visualization tools. In particular, we enhanced the differential analysis tools, which now enable online automated identification of DNA methylation abnormalities in human disease in a case-control or disease–disease manner. To facilitate further mining of the disease methylome, three new web tools were developed for cluster analysis, functional annotation and survival analysis. DiseaseMeth version 2.0 should be a useful resource platform for further understanding the molecular mechanisms of human diseases.
INTRODUCTION
DNA methylation is a chemical modification on DNA sequence (1) that directs and restricts cell differentiation during growth and development. Because of its important role in mammals, the abnormal occurrence or removal of methylation is likely to contribute to the risk of diseases. Therefore, DNA methylation associated with disease has been a focus of extensive research interest. To provide a comprehensive DNA methylation repository of human diseases, we developed DiseaseMeth (2), which is a web-based resource of aberrant methylomes related to human diseases. DiseaseMeth included 175 large-scale disease methylation datasets from 88 diseases produced by 19 techniques, as well as over 14 000 items of curated experimental information mined from PubMed. In the five years since its publication, DiseaseMeth has been used in disease methylation analyses by more than 7000 visitors from more than 90 countries.
Recently, several high-throughput technologies have been developed for detection of the DNA methylome in a large number of samples including human diseases. The Illumina Infinium Human Methylation450BeadChip has been widely used to profile DNA methylation in various human diseases, including a large number of cancer samples from The Cancer Genome Atlas (TCGA) project (3). TCGA project, for example, used an Infinium BeadChip to map methylation profiles in more than 10 000 samples from 32 cancer types including glioblastoma, breast cancer and papillary kidney carcinoma (4–6). In addition, whole-genome bisulfite sequencing (WGBS) has been used to detect whole methylomes in human diseases at single-site resolution. By analyzing WGBS datasets, Hovestadt et al. (7) found that alterations in DNA methylation of novel candidate genes (e.g. LIN28B) in medullablastoma resulted in alternative promoter usage and/or differential messenger RNA/microRNA expression. Reduced representative bisulfite sequencing(RRBS) is another bisulfite-based technology that is suitable for obtaining methylation information from regions of high CpG content (e.g. CpG islands) and has been used to provide detailed epigenetic status in various human disease tissues and cell lines (8,9). For example, Hascher et al. used RRBS to map the genome-wide methylation landscape in lung cancer and found that DNA methylation had an influence on poly comb target genes (10).
DNA methylation experimental technologies have helped to uncover more and more differential DNA methylation of genes in disease. Agarwal et al. found that DNA-methyl transferase 1 (DNMT1) was overexpressed in many cancers and was correlated to aberrant methylation in nasopharyngeal carcinoma. It may be the risk stratification and final outcomes in patients with nasopharyngeal carcinomas (11). Lan et al. (12) found again in osuccinate synthase 1 (ASS1) deficiency maybe linked to therapeutic sensitivity to arginine-depriving agents and promote tumor aggressiveness through its newly identified tumor suppressor function. Andres et al. (13) confirmed global DNA hypomethylation in rheumatoid arthritis patients, with specificity for some blood cell subpopulations. Hardy et al. (14) found differential DNA methylation of the PPARγ promoter in 26 patients with biopsy-proven non-alcoholic fatty liver disease. Moreover, based on the rapidly increasing amount of data, a number of relationships between DNA methylation of genes and diseases have been reported. For example, in prostate cancers, APC, MGMT and RASSF1A were methylated in at least 60% of the patients in the study, while other genes, including COX-2, DAPK1, CDH1, CDKN2A, RUNX3 and THBS1 were methylated at frequencies lower than 35% (15). Further, it was suggested that high levels of CpG island hypermethylation might serve as a potential biological marker for aggressive prostate cancer.
Large amounts of methylation data are now available from various sources including TCGA project, the International Human Epigenome Consortium (IHEC, http://ihec-epigenomes.org/) and the Gene Expression Omnibus (GEO) (16). Databases, including DiseaseMeth (2), MethHC (17), MethyCancer (18), NGSmethDB (19) and MethBase (20), have been developed to store this methylation data and disease information. However, an online platform that integrates high-throughput methylome data and online analyses is still needed for detecting disease–gene associations. To meet this need, we have updated DiseaseMeth and built an online platform for data integration and analyses.
In this paper, we present DiseaseMeth version 2.0, which represents a qualitative leap from the previous version (2). DiseaseMeth version 2.0 is focused not only on curated information about diseases, genes and corresponding methylation data, but also on predicted associations between diseases of interest and methylation of specific DNA regions based on the vast amounts of data that it contains. DiseaseMeth version 2.0 contains methylation data of 32 701 samples from 88 diseases together with 679 602 associations between diseases and methylation of genes. To facilitate research on the disease methylome, DiseaseMeth version 2.0 has been updated and several new tools and features have been added as follows. (i) The online workflow in the analysis page was developed for differential DNA methylation analysis in selected diseases and further interpretation for the results, including cluster analysis, functional annotation, survival analysis and searching the existing disease–gene associations stored in this database. (ii) The search engine has been enhanced to search DNA methylation data in selected samples or in given genomic regions. The search results can be visualized and users can select data for differential DNA methylation analysis. (iii) Four categories of associations between diseases and genes are revealed according to the text mining. (iv) We rebuild the genome browser, DisMethBrowser, which contains abundant annotation tracks with supporting customized views in the genomic scale.
In summary, DiseaseMeth version 2.0 not only enlarges the data of increased DNA methylation, but also provides new tools to explore the relationships between methylation of genes and diseases. DiseaseMeth version 2.0 is a comprehensive resource that will help researchers understand diseases from an epigenetics viewpoint.
DATA EXPANSION AND PRE-PROCESSING
DiseaseMeth version 2.0 is updated to include the increased amount of DNA methylation data and associations between diseases and genes (Table 1). There are 88 kinds of disease in the updated version. Among them, 74 diseases were detected by high-throughput experimental technology including 32 701 methylation profiles. The data sets were collected from huge international disease projects including TCGA and public genome databases including GEO (16).
Table 1. Expanded data content in DiseaseMeth version 2.0 compared with the previous version of DiseaseMeth.
| Total No. | Items | No. of each item | |
|---|---|---|---|
| Illumina Infinium HumanMethylation450 BeadChip | 16 317 (16 317) | ||
| Illumina Infinium HumanMethylation27 BeadChip | 11 295 (9506) | ||
| Reduced Representation Bisulfite Sequencing | 88 (30) | ||
| Methylation profiles in human diseases | 32 701 (28 471) | Whole-Genome Bisulfite Sequencing | 80 (80) |
| Methylated DNA Immunoprecipitation Sequencing | 58 (58) | ||
| Methylation-sensitive Restriction Enzyme Sequencing | 33 (33) | ||
| Illumina GoldenGate DNA methylation Beadchip | 1280 (1265) | ||
| Other | 3550 (1182) | ||
| Disease and Gene Association | 679 602 (471 916) | Inferred relationship | 256 919 (42 835) |
| Potential relationship | 429 081 (429 081) | ||
| experimentally verified | 2880 (535) | ||
| Transcription Factor Binding Sites | 438 044 (438 044) | Transcription Factor Binding Sites | 438 044 (438 044) |
| DNase I hypersensitive sites | 1 867 665 (1 867 665) | DNase I hypersensitive sites | 1 867 665 (1 867 665) |
| Genome information | 99 865 (0) | UCSC mRNA | 58 939 (0) |
| CpG Island | 40 926 (0) |
The numbers in parentheses indicate the expanded data in DiseaseMeth version 2.0.
For ease of usage and to reveal further information, we processed and standardized the data sets. From TCGA project, we downloaded 9795 profiles of the Illumina Infinium HumanMethylation450 BeadChip, 2728 profiles of the Illumina Infinium HumanMethylation27 BeadChip and 47 profiles of whole-genome bisulfite sequencing. The Illumina Infinium HumanMethylation27 BeadChip data were converted into the UCSC assembly (21) of the February 2009 human reference sequence (GRCh37) with LiftOver (21). In addition, we downloaded 6522 profiles of Illumina Infinium HumanMethylation450 BeadChip, 8567 profiles of Illumina Infinium HumanMethylation27 BeadChip and 88 profiles of RRBS from GEO. We converted the methylation levels represented by M-values to β-values (22). For the raw sequencing data from bisulfite sequencing platforms that we downloaded from the sequence read archive (23), we performed read mapping and methylation calling using the bisulfite mapping tool Bismark (24) to obtain the methylation levels. The source information about disease status, sample ID, research ID or study ID, data platform, download links and tissue or cell line of each methylation profile was assembled and stored into a MySQL backend database. Both the disease methylation profiles and the query/analysis results could be downloaded from DiseaseMeth version 2.0 for the use of academic research.
Moreover, in DiseaseMeth version 2.0, the associations between diseases and methylation of genes were increased to 679 602 items including 256 919 that were predicted from the 32 701 profiles. To provide distinct information, the associations between diseases and genes were classified into four groups: 2880 experimentally verified disease–gene association; 214 084 inferred DNA methylation mediated disease–gene associations identified by computational methods from the reported analysis of DNA methylation profiles; and 429 081 potential methylation-associated disease–gene associations that were published in DisGeNET (25) which integrates information on disease–gene associations from several public data sources and the literature. In addition, based on the DNA methylation profiles hosted in DiseaseMeth version 2.0, we identified 42 835 other disease–gene associations by enhanced t-test (see Supplementary Methods) with restriction of P < 0.01 and absolute difference of methylation > 0.2, which has been integrated into the enhanced DNA methylation analysis toolkit in this update. DiseaseMeth version 2.0 provides information about disease–gene associations, including disease name, disease ID, gene name, groups of the association, evidence and origin.
UPDATE OF THE DISEASEMETH CORE
Updated toolkit for analyses of differential DNA methylation and relationships between gene and disease
The differential DNA methylation analysis tool was updated by integrating the Student's t-test, Shannon entropy and the edgeR (26), minfi (27) and samr (28) algorithms to analyze differences across two or more groups of human disease data in a case-control or disease–disease manner. In order to recover the significantly differential methylated genes/CpG sites, the measure of significant P-values and absolute difference of the mean methylation levels have been considered simultaneously in our method (see Supplementary Methods). In addition, differential methylation can be performed in the seven RefSeq gene-related regions or the CGI-related regions according to the interesting.
Disease–gene associations were defined as significantly differential methylated genes among case-control groups of a particular disease. For reference, the currently reported disease–gene associations detected by experimental methods, computational methods or integrated strategies inferred associations are listed in DiseaseMeth version 2.0. Some basic information including mean methylation, P-value, the gene expression regulation, disease progression and the methylation type have been provided for future studies. Disease–disease and gene–gene association analyses were also integrated into the above analysis flow through a correlation analysis strategy.
DisMethBrowser: newly developed disease methylome visualization tool
We developed a new disease methylome browser called DisMethBrowser to provide a user-friendly interface. We increased the annotation tracks with transcriptional elements from UCSC. In DisMethBrowser, the configuration of the track option lists was further simplified so that users can easily obtain the track data of interest through the search engine by clicking the ‘Add Samples’ button.
NEW TOOLS FOR EXTENDED ANALYSIS OF DIFFERENTIAL METHYLATED GENES
To explore the regulation mechanism of methylation causing disease, three new web tools were developed to uncover information that may be hidden in the big data, including cluster analysis, functional annotation and survival analysis. The cluster analysis tool exhibits the characteristics of the methylation profile in disease samples within input genes and provides a heatmap of the results. These results can be used to directly visualize the methylation profile and compare the methylation levels between cases and controls. The extended functional annotation tool offers convenient access to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (29) and the Genomic Regions Enrichment of Annotations Tool (GREAT) (30), which can be used to identify the functions of differential genes or regions. For genes or transcripts, a functional analysis can be performed through the DAVID web service, while, for genomic intervals, GREAT can be used instead. Similarly, a gene of interest could be submitted to PROGgene (31) for survival analysis of a chosen disease to confirm whether or not a gene is related to the survival time of patients with a specific cancer.
DATABASE USE AND ACCESS
Search tools provided by DiseaseMeth version 2.0
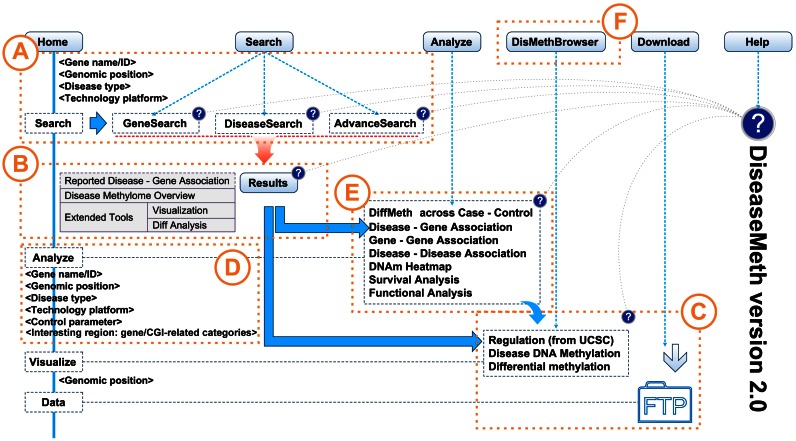
DiseaseMeth version 2.0 can be accessed freely at http://bioinfo.hrbmu.edu.cn/diseasemeth/. The search module in DiseaseMeth version 2.0 provides three query types, GeneSearch, DiseaseSearch and AdvanceSearch (Figure 1A). On the GeneSearch page, users can specify a Gene symbol (gene name/transcript ID) or Genomic position to obtain the methylation levels of genes across disease samples. On the DiseaseSearch page, users can select the Disease type to obtain a heatmap of the methylation landscape for the selected disease. The AdvanceSearch page allows users to specify a more accurate query. By inputting Gene name/Transcript ID or Genomic position, and selecting Disease type and Technology platform, users can obtain a more specific data set. In the result page, users can select samples for further analyses like visualization and differential DNA methylation status analysis (Figure 1B). For the differential methylation analysis, besides the Student′s t-test, a number of other statistical methods have been built-in to DiseaseMeth version 2.0, such as QDMR, a method for quantitative identification of differentially methylated region (32) based on Shannon entropy, edgeR, minfi and samr. Following a differential analysis, users can make use of the build-in tools to analyze the results, including visualization of the results in the genome browser, functional annotation and downloading the result information or the origin methylome data (Figure 1C).
Figure 1.
Content and construction of DiseaseMeth version 2.0. (A) Search engine options: GeneSearch, DiseaseSearch and AdvanceSearch. (B) Search results include information of Disease–Gene Association and Disease Methylome Overview, as well as a link to Extended Tools. (C) DisMethBrowser and Data downloads. (D) Setting options for the Differential DNA Methylation Analysis Tool. (E) Seven tabs used for the analysis results. (F) DisMethBrowser of DiseaseMeth version 2.0 for the visualization of methylation landscape of a specified region.
Online tools for differential methylation identification and functional analysis
Users can input a group of gene symbols, transcript IDs, or genomic intervals in the search table to analyze both their associations with selected diseases and the functional correlations between the input genomic regions (Figure 1D). When gene symbols or transcript IDs are provided, the default analysis regions are the promoter regions defined as from 2000 bp upstream of the transcription start site (TSS) to 500 bp downstream of the TSS of RefSeq transcripts (-2∼0.5 kb). Besides, four other definitions of the promoter region, including -2, -1.3∼0.2, -1∼0.5 and -1.5∼0.5 kb, have been provided for users to refer to. However, users can customize the length of the promoter regions to be analyzed. Differential DNA methylation status can also be performed in other regions of the transcripts by selecting ‘Region of Interest’ to access gene body, 5′UTR, 3′UTR, exon, intron, down2kb and up2kb, or CpG region divide to access CpG island, CpG island shore and CpG island shelves.
The results are divided into seven tabs (Figure 1E): Diff Case-Control, Disease–Gene association, Methylation Profile Cluster, Survival Analysis for Cancer, Functional Annotation, Gene-Gene Association and Disease–Disease Association. The Diff Case-Control tab displays the result of a differential DNA methylation status analysis using the method selected by the users for each keyword between the case and control for each selected disease. Users can obtain a box plot for each keyword to visualize the results of the differential analysis. The Disease-Gene association tab detects stored associations between the input genomic regions and the selected diseases. The Methylation Profile Cluster tab provides a heatmap that displays the methylation profile of samples and the cluster results for each of the selected diseases. The Survival Analysis for Cancer tab provides a Kaplan-Meier curve for each selected gene. The Functional Annotation tab provides the annotations of the selected genes using DAVID v6.7. These analysis tools allow users to mine large amounts of data to determine if a given genomic region is related to the selected diseases.
DisMethBrowser of DiseaseMeth version 2.0
DisMethBrowser (Figure 1F) can be used to visualize the methylation data of samples or the results of differential methylation analysis. Methylation data in the various samples can be added via the search page and the annotation tracks can be controlled in the DisMethBrowser. Genes symbol, transcript ID or genomic interval can be submitted to browse the methylation level and the annotation of a given region. DisMethBrowser with the annotation tracks allow users to easily browse and understand the methylation data and the results of differential analysis.
A case application of DiseaseMeth version 2.0 in a systematic analysis for selected carcinomas
On the ‘AdvanceSearch’ page, we input NKAPL as the Gene Symbol, selected Rectum adenocarcinoma[READ], Liver hepatocellular carcinoma [LIHC], Prostate adenocarcinoma [PRAD] and Breast invasive carcinoma[BRCA] as the Disease, and chose 450k (Illumina Infinium HumanMethylation450 BeadChip) as the Technology Experimental Platform (Figure 2A). After clicking the ‘Search’ button at the bottom of the page and further chose ‘promoter’ with the default parameters ‘2000 bp upstream of TSS to 500 bp downstream’ to continue, then we obtained the search result page (Figure 2B), which listed one entries of the association between disease and gene, and 2153 entries of DNA methylomes that are stored in DiseaseMeth version 2.0. It showed that NKAPL was significantly differential methylated in BRCA (t-test, P < 0.01), the mean methylation level in cancer samples was 0.621 and the mean methylation level in normal samples was 0.367. The methylation profile indicated that it was hyper-methylated in breast cancer. By submitting an analysis request through the ‘Extended Tools’ on the bottom of the search result page or through the Analyze menu options (Figure 2C), we performed the differential methylation analysis for the four selected carcinomas. Then it displayed the transcription matching results for each gene symbol (OR2M3, PRDM14, MIR663A, NKAPL and OR10K2) on the top of the page. Here, we select the gene promoter by checking ‘2 kb upstream of TSS to 0.5 kb downstream’ as the ‘Region of Interest’ for the following analysis (Figure 2D). The results given in the Diff Case-Control tab (Figure 2F) show that all five input genes, MIR663A, OR10K2, NKAPL, PRDM14 and OR2M3, were significantly differential methylated in BRCA. By clicking the ‘+/-’ button, a boxplot for the corresponding gene was displayed to visualize methylation levels between case-control samples. The Disease–Gene Association tab (Figure 2G) listed 22 reported records, which show that these five genes were also identified in other researches. Clicking the Methylation Profile tab (Figure 2H) displayed a heatmap view of the whole methylation landscape of the five genes in the specified disease. The heatmap view could be switched by clicking the tab-button at the top of the panel. The Survival Analysis tab (Figure 2I) provided a Kaplan–Meier curve for the selected gene NKAPL, which showed that the BRCA patients were divided into high-risk and low-risk groups based on the expression of NKAPL and the survival times of the two groups were significantly different (t-test, p = 0.0036). Through the functional annotations (Figure 2J), we found that the five genes were significantly enriched in the Olfactory transduction pathway. The relationship analysis based on DNA methylation of the five genes (Figure 2K) revealed that NKAPL was highly relevant to the microRNA coding gene MIR663A and PRDM14, and that PRADM14 was highly relevant to MIR663A. For the Disease–Disease Relationship Analysis (Figure 2L), we found that BRCA was highly related to READ and LIHC at the methylation level of the five submitted genes. Finally, we created the methylation landscape for NKAPL in the DisMethBrowser (Figure 2M).
Figure 2.
A case study showing the usage of DiseaseMeth version 2.0. (A) The AdvanceSearch page with NKAPL input as the Gene Symbol, READ, LIHC, PRAD and BRCA selected as Disease, and 450k chosen as the Technology Experimental Platform. (B) Search results page. (C) The Analyze menu options page. (D) Transcription matching results for each gene symbol on the top of the page. And the ‘Select Region of Interest’ option controller. (E) The analysis results included in seven tabs. (F) The Diff Case-Control tab. (G) The Disease–Gene Association tab. (H) The Methylation Profile tab. (I) The Survival Analysis tab. (J) The functional annotation results tab. (K) Relationship analysis based on DNA methylation of genes. (L) The Disease–Disease Relationship Analysis tab. (M) Visualization of NKAPL in DisMethBrowser.
Comparison of DiseaseMeth version 2.0 with other databases
To highlight the current advantages of DiseaseMeth version 2.0, we compared DiseaseMeth version 2.0 with other related 15 databases based on a total of 44 sub-characters about concerned 13 features. Detailed results of the comparisons are available in Supplementary Table S1. For example, the data records in PubMeth (33), DDMGD (34) and MeInfoText 2.0 (35) were automatically compiled via text mining, only PubMeth processed a following manual curation. Additionally, MeInfoText 2.0, MethDB (36), MethyCancer, PubMeth, MENT (37), CMS (38), PCMdb (39), PD_NGSAtlas (40) and MethmiRbase (41) focused on human cancers/disorders, while MethylomeDB (42) is focused on the brain tissues. MeT-DB (43) is a comprehensive database for N6-methyladenosine (m6A) mammalian methyltranscriptome. DiseaseMeth version 2.0 collected nearly all existed kinds of methylation datasets from different technology platforms. Totally, 32 701 high-throughput DNA methylation profiles of 74 kinds of human diseases have been assembly released in this updated version. Besides, we included 2880 single-gene methylation appraisal analysis by manual curation. The main differential of DiseaseMeth version 2.0 with other databases is the tools for online identification and analysis of disease–gene associations based on high-throughput DNA methylation profiles. Most of those databases only included information on methylated genes in the diseases. The online analysis tools will help to provide more and more information about diseases and genes. In summary, DiseaseMeth version 2 is a synthetical platform gathering with the huge data storing and analyzing of methylation profiles in human diseases.
FUTURE DEVELOPMENT
The importance of epigenetics now is widely recognized, especially in the development and occurrence of diseases. DiseaseMeth version 2.0 incorporates curated information about methylation-associated disease genes and detailed analyses of associations between diseases and genes using high-through put methylation data. These improved tools are convenient for users to query, analyze and understand the analysis results without the need for any other external tools. What is more, we will continuously enhance the function of DiseaseMeth. To build a comprehensive DNA methylation database of human diseases, data from different diseases and platforms will be added to keep DiseaseMeth up-to-date. Additional data from various omics studies will also be incorporated to contribute to the comprehensive analysis of methylation data. Through our efforts, we expect that DiseaseMeth will continue to contribute to research into further understanding of epigenetic regulation mechanisms and even toward the epigenetic diagnosis and treatment of human diseases.
Acknowledgments
We thank TCGA, GEO, the NIH Roadmap Epigenomics, ENCODE and other projects for generating and sharing the data used in this paper. We acknowledge Wenhua Lv, Xinyu Wang, Libo Wang, Ce Ci, Yan Huang, Shipeng Shang, Min Zhang, Mengying Zhang and Zixu Wang for their valuable suggestion and discussion on database update.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31371334, 61403112, 81573021, 61402139]; Natural Science Foundation of Heilongjiang Province [ZD2015003]; Innovation and Technology Special Fund for researchers of Harbin Science and Technology Bureau [2015RAXXJ052]; Innovation Research Fund for Graduate Students of Harbin Medical University [YJSCX2014-23HYD, YJSCX2014-24HYD]. Funding for open access charge: National Natural Science Foundation of China [31371334 and 81573021].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Lv J., Liu H., Su J., Wu X., Liu H., Li B., Xiao X., Wang F., Wu Q., Zhang Y. DiseaseMeth: a human disease methylation database. Nucleic Acids Res. 2012;40:D1030–D1035. doi: 10.1093/nar/gkr1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C., et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research, N. Linehan W.M., Spellman P.T., Ricketts C.J., Creighton C.J., Fei S.S., Davis C., Wheeler D.A., Murray B.A., Schmidt L., et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hovestadt V., Jones D.T., Picelli S., Wang W., Kool M., Northcott P.A., Sultan M., Stachurski K., Ryzhova M., Warnatz H.J., et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 8.Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H., et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hascher A., Haase A.K., Hebestreit K., Rohde C., Klein H.U., Rius M., Jungen D., Witten A., Stoll M., Schulze I., et al. DNA methyltransferase inhibition reverses epigenetically embedded phenotypes in lung cancer preferentially affecting polycomb target genes. Clin. Cancer Res. 2014;20:814–826. doi: 10.1158/1078-0432.CCR-13-1483. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal S., Amin K.S., Jagadeesh S., Baishay G., Rao P.G., Barua N.C., Bhattacharya S., Banerjee P.P. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol. Cancer. 2013;12:99–111. doi: 10.1186/1476-4598-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J., Tai H.C., Lee S.W., Chen T.J., Huang H.Y., Li C.F. Deficiency in expression and epigenetic DNA Methylation of ASS1 gene in nasopharyngeal carcinoma: negative prognostic impact and therapeutic relevance. Tumour Biol. 2014;35:161–169. doi: 10.1007/s13277-013-1020-8. [DOI] [PubMed] [Google Scholar]
- 13.de Andres M.C., Perez-Pampin E., Calaza M., Santaclara F.J., Ortea I., Gomez-Reino J.J., Gonzalez A. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Res. Ther. 2015;17:233–242. doi: 10.1186/s13075-015-0748-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy T., Zeybel M., Day C.P., Dipper C., Masson S., McPherson S., Henderson E., Tiniakos D., White S., French J., et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2016;112:E5503–E5512. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang G.H., Lee S., Lee H.J., Hwang K.S. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J. Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 16.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W.Y., Hsu S.D., Huang H.Y., Sun Y.M., Chou C.H., Weng S.L., Huang H.D. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X., Chang S., Zhang J., Zhao Q., Xiang H., Kusonmano K., Yang L., Sun Z.S., Yang H., Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisen S., Barturen G., Alganza A.M., Hackenberg M., Oliver J.L. NGSmethDB: an updated genome resource for high quality, single-cytosine resolution methylomes. Nucleic Acids Res. 2014;42:D53–D59. doi: 10.1093/nar/gkt1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Q., Decato B., Hong E.E., Zhou M., Fang F., Qu J., Garvin T., Kessler M., Zhou J., Smith A.D. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One. 2013;8:e81148. doi: 10.1371/journal.pone.0081148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587–596. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database, C. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer-Mehren A., Rautschka M., Sanz F., Furlong L.I. DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics. 2010;26:2924–2926. doi: 10.1093/bioinformatics/btq538. [DOI] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goswami C.P., Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J. Clin. Bioinform. 2013;3:22–31. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Liu H., Lv J., Xiao X., Zhu J., Liu X., Su J., Li X., Wu Q., Wang F., et al. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res. 2011;39:e58. doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ongenaert M., Van Neste L., De Meyer T., Menschaert G., Bekaert S., Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bin Raies A., Mansour H., Incitti R., Bajic V.B. DDMGD: the database of text-mined associations between genes methylated in diseases from different species. Nucleic Acids Res. 2015;43:D879–D886. doi: 10.1093/nar/gku1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang Y.-C., Lai P.-T., Dai H.-J., Hsu W.-L. MeInfoText 2.0: gene methylation and cancer relation extraction from biomedical literature. BMC Bioinformatics. 2011;12:471–479. doi: 10.1186/1471-2105-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amoreira C., Hindermann W., Grunau C. An improved version of the DNA Methylation database (MethDB) Nucleic Acids Res. 2003;31:75–77. doi: 10.1093/nar/gkg093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baek S.J., Yang S., Kang T.W., Park S.M., Kim Y.S., Kim S.Y. MENT: methylation and expression database of normal and tumor tissues. Gene. 2013;518:194–200. doi: 10.1016/j.gene.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 38.Gu F., Doderer M.S., Huang Y.W., Roa J.C., Goodfellow P.J., Kizer E.L., Huang T.H., Chen Y. CMS: a web-based system for visualization and analysis of genome-wide methylation data of human cancers. PLoS One. 2013;8:e60980. doi: 10.1371/journal.pone.0060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagpal G., Sharma M., Kumar S., Chaudhary K., Gupta S., Gautam A., Raghava G.P. PCMdb: pancreatic cancer methylation database. Sci. Rep. 2014;4:4197. doi: 10.1038/srep04197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z., Li Y., Chen H., Lu J., Thompson P.M., Chen J., Wang Z., Xu J., Xu C., Li X. PD_NGSAtlas: a reference database combining next-generation sequencing epigenomic and transcriptomic data for psychiatric disorders. BMC Med. Genomics. 2014;7:71–82. doi: 10.1186/s12920-014-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agustriawan D., Wijaya E.B., Huang C.-H., Lim E., Hsueh I.-C., Kurubanjerdjit N., Tzeng K.-R., Ng K.-L. MethmiRbase: a database of DNA methylation and miRNA expression in human cancer. Lecture Notes in Comp Sci. 2016;2221:73–76. [Google Scholar]
- 42.Xin Y., Chanrion B., O'Donnell A.H., Milekic M., Costa R., Ge Y., Haghighi F.G. MethylomeDB: a database of DNA methylation profiles of the brain. Nucleic Acids Res. 2012;40:D1245–D1249. doi: 10.1093/nar/gkr1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Flores M.A., Meng J., Zhang L., Zhao X., Rao M.K., Chen Y., Huang Y. MeT-DB: a database of transcriptome methylation in mammalian cells. Nucleic Acids Res. 2014;43:D197–D203. doi: 10.1093/nar/gku1024. [DOI] [PMC free article] [PubMed] [Google Scholar]