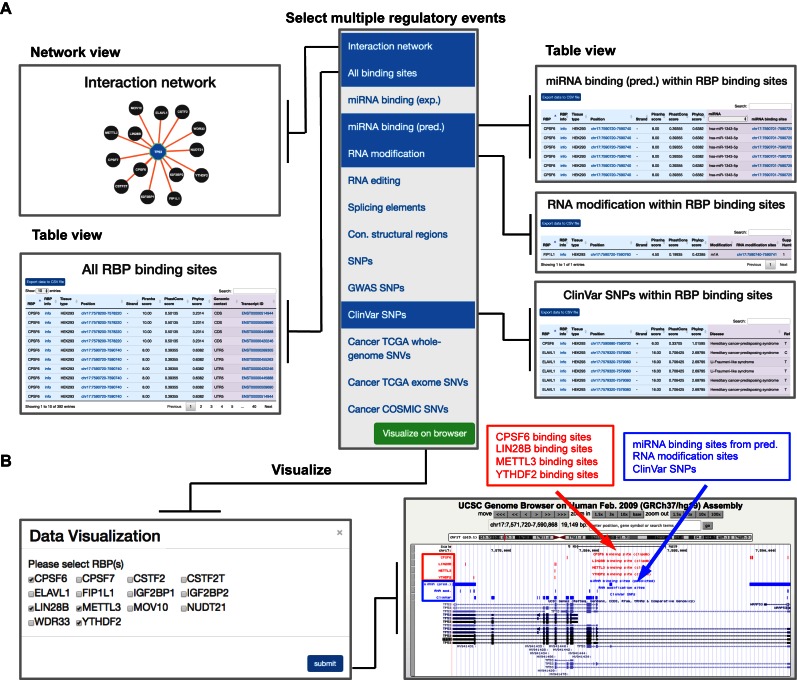
Figure 3.
‘POSTAR’ search enables integrative viewing of multiple RBP binding sites and their potential to post-transcriptionally regulate a target gene (TP53 as an example). (A) In the PAR-CLIP Piranha data module, users may select ‘interaction network’, ‘all binding sites’, and multiple regulatory elements, including ‘miRNA binding (pred.)’, ‘RNA modification’, and ‘ClinVar SNPs’, to obtain detailed information in one page. (B) By clicking on the ‘Visualize in browser’ button (green), a user can select four RBPs among all bound RBPs to simultaneously visualize their binding sites (red tracks) and regulatory events (blue tracks) in an integrative manner via the UCSC genome browser.