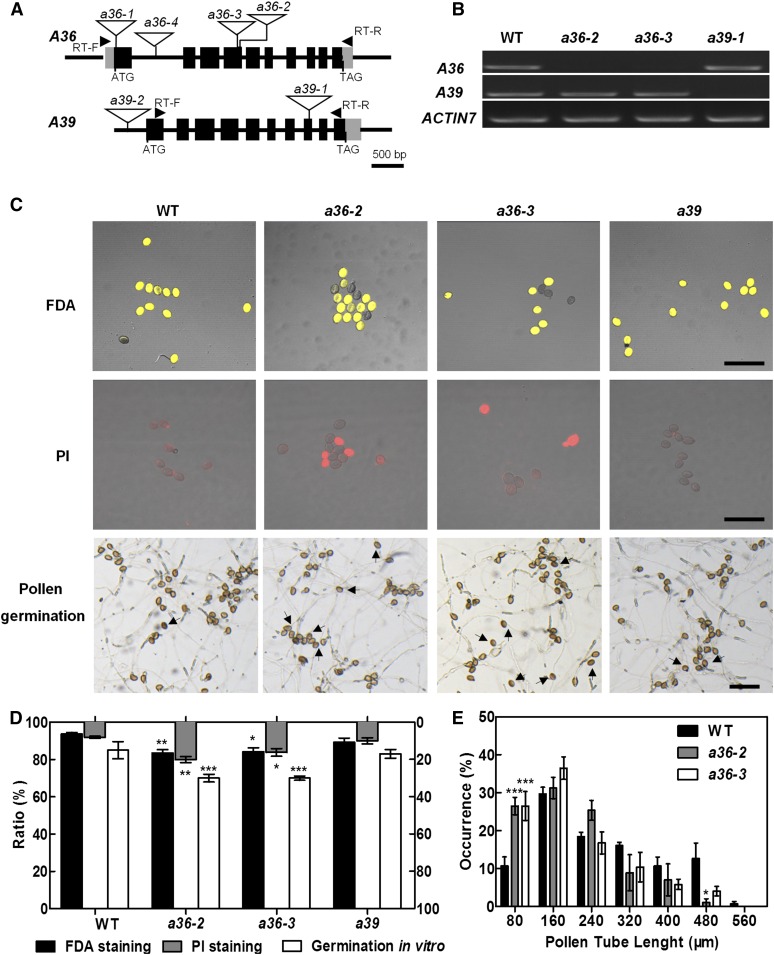
Figure 2.
a36 displays reduced pollen grain activity and pollen germination ratio in vitro. A, Genomic organization of the intronless A36 and A39 genes and positions of a36-2, a36-3, and a39-1 T-DNA insertions. The positions of the primers used for RT-PCR are indicated by arrowheads. Exons are represented by black boxes and introns by lines. The locations of the T-DNA insertions are indicated with triangles. B, RT-PCR analysis from open-flower cDNAs shows no A36 transcripts in a36-2 and a36-3 and no A39 transcripts in a39-1. ACTIN7 was used as an internal control. Amplification was performed for 25 cycles for ACTIN7 and for 30 cycles for A36 (1,206 bp) and A39 (1,258 bp). C, Pollen viability of the wild type (WT), a36-2, a36-3, and a39 by FDA (top row) and PI (middle row) staining. FDA-positive (viable) and PI-positive (unviable) pollen are indicated in yellow and red, respectively. The bottom row shows the pollen germination assay in vitro for 8 h. Pollen of the wild type and a39 germinated very well and produced actively growing pollen tubes. By contrast, pollen germination in a36-2 and a36-3 was impaired. Arrows designate pollen that did not germinate. Bars = 100 μm. D, Statistical analysis of pollen viability by FDA and PI staining and germination rates of pollen grains from wild-type, a36-2, a36-3, and a39 plants in vitro. Data were collected from three independent experiments (n > 900). E, Statistical analysis of pollen tube length distribution after incubation for 4 h in vitro. Data were collected from three independent experiments (n = 60). For D and E, error bars show se and asterisks indicate values that differ significantly from those of the wild type (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, calculated using two-way ANOVA).