The crystal structure of a rice CesA8 plant-conserved region (P-CR), solved to 2.4 Å resolution, is dockedto a model of the catalytic domain of CesA8 using SAXS molecular envelopes.
Abstract
The crystallographic structure of a rice (Oryza sativa) cellulose synthase, OsCesA8, plant-conserved region (P-CR), one of two unique domains in the catalytic domain of plant CesAs, was solved to 2.4 Å resolution. Two antiparallel α-helices form a coiled-coil domain linked by a large extended connector loop containing a conserved trio of aromatic residues. The P-CR structure was fit into a molecular envelope for the P-CR domain derived from small-angle X-ray scattering data. The P-CR structure and molecular envelope, combined with a homology-based chain trace of the CesA8 catalytic core, were modeled into a previously determined CesA8 small-angle X-ray scattering molecular envelope to produce a detailed topological model of the CesA8 catalytic domain. The predicted position for the P-CR domain from the molecular docking models places the P-CR connector loop into a hydrophobic pocket of the catalytic core, with the coiled-coil aligned near the entrance of the substrate UDP-glucose into the active site. In this configuration, the P-CR coiled-coil alone is unlikely to regulate substrate access to the active site, but it could interact with other domains of CesA, accessory proteins, or other CesA catalytic domains to control substrate delivery.
Plant cellulose microfibrils are synthesized at the plasma membrane surface by large, oligomeric protein complexes (cellulose synthase complexes [CSCs]) arranged in hexagonal arrays called particle rosettes (Giddings et al., 1980; Mueller and Brown, 1980). Individual plant cellulose synthases (CesAs) in these complexes each catalyze the synthesis of a single (1→4)-β-d-glucan chain, which is extruded through membrane channels and coalesces with multiple other chains to form the para-crystalline cellulose microfibril at the plasma membrane surface.
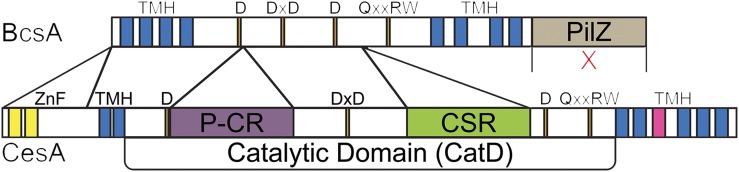
Plant cellulose synthases evolved from prokaryotic ancestral genes (Nobles et al., 2001). Bacterial synthases make single chains of (1→4)-β-d-glucan that are guided through the periplasm by accessory proteins and are integrated at the peripheral membrane into crystalline forms secreted into the medium as biofilms or pellicles (Kondo et al., 2002). Bacterial cellulose synthases were first discovered in Gluconacetobacter xylinus (syn. Acetobacter xylinum; Wong et al., 1990; Saxena et al., 1990). Four catalytic signature sequences containing D, DxD, D, and Q/RxxRW residues considered essential for substrate binding and catalysis in these synthases are shared by several related nucleotide-sugar transferases (Saxena et al., 1995; Fig. 1). These four broadly conserved catalytic sequences were used to identify putative plant cellulose synthases (Pear et al., 1996).
Figure 1.
Bar diagram of the domains and signature sequences of cellulose synthases in bacteria (BcsA) and plants (CesA). Like BcsA, CesA possesses four catalytic signature motifs (orange) containing conserved D, DxD, D, and Q/RxxRW residues. Plants differ from bacterial synthases by the absence of the PilZ activator domain (beige) and the addition of three unique domains, a RING-type zinc finger domain (ZnF; yellow) in the extended N terminus and the P-CR (purple) and CSR (green) domains inserted into the CatD. Recent work has indicated that CesA also may differ from BcsA in its number of transmembrane helices (TMH; blue), with BcsA possessing eight transmembrane helices and CesA likely possessing seven transmembrane helices, with the putative TMH5 (pink) hypothesized to occupy an interfacial orientation along the membrane (Slabaugh et al., 2016).
The characterization of the crystal structure of the bacterial cellulose synthase (BcsA) complex from Rhodoobacter sphaeroides was a major breakthrough in understanding the catalytic mechanism for sugar transfer, chain elongation, and product channeling and assigned a molecular function to the signature sequences (Morgan et al., 2013). The initial D and DxD signature sequences bind the UDP substrate at the uracil and diphosphate, respectively. The next D is the catalytic base oriented in a position to access the end of the glucan chain, and the Q/RxxRW signature sequence forms a major part of the binding site for the terminal disaccharide. The catalytic core structure of BcsA, a variant of the classical nucleotide-binding fold (Rossmann et al., 1974), is conserved among several similar types of nucleotide-sugar transferases, including CesAs. However, plant CesAs are distinguished from their bacterial homologs by the presence of three unique domains, one forming the extended N terminus and two located in the catalytic core. The bacterial C-terminal PilZ domain that is responsible for synthesis activation via cyclic di-GMP is absent in plant CesAs (Fig. 1). The novel plant-specific domains might replace the regulatory function of the PilZ or serve in the assembly of CesA isoforms into oligomeric complexes, a requirement that distinguishes bacterial and plant cellulose synthases. For example, the N-terminal addition of the cytosolic RING-type zinc finger domain was shown to function in redox-dependent dimerization and is thought to recruit CesAs into complexes (Kurek et al., 2002).
The functions of the two domains within the plant CesA catalytic domain (CatD), the plant-conserved region (P-CR) and the class-specific region (CSR), have not been fully defined. An examination of the sequences of the P-CR and CSR indicates no significant similarity to any domain of known function. Straightforward sequence comparisons show that the P-CR domain is highly conserved among all CesA isoforms of flowering plants. The sequences of CSR domains, first thought to be hypervariable (Pear et al.,1996; Delmer, 1999), form a multiclade subgroup structure that is conserved in CesAs across plant species (Vergara and Carpita, 2001; Penning et al., 2009). The multigene families of plant CesAs (Richmond and Somerville, 2000) are expressed differentially during plant growth; at least three isoforms are expressed during primary wall formation, and a different set of three isoforms is expressed during secondary cell wall synthesis (Tanaka et al., 2003; Appenzeller et al., 2004; Brown et al., 2005). The characterization of mutants for each of these isoforms indicates that they are nonredundant and perform specific roles within the CSC (Taylor, 2008). The sequence variation of the CSRs of these isoforms constitutes the primary distinction among the CesAs, defining the classes that are expressed in these groups. Recently, the CSR was shown to control redox- and concentration-dependent dimerization of rice (Oryza sativa) CesA8 CatDs in vitro (Olek et al., 2014), which may indicate how this domain functions in the CSC assembly.
In contrast, the P-CR has not been definitively shown to have a distinct function in CSC assembly or in either (1→4)-β-d-glucan chain or microfibril synthesis. The catalytic core domain homologous to bacterial sequences is predicted to adopt the same nucleotide-binding fold as seen in the BcsA structure (Olek et al., 2014). However, the P-CR and CSR domains lack significant similarity to any of the known structures in the Protein Data Bank (PDB; Berman et al., 2000) and, consequently, also lack SCOP (Murzin et al., 1995), CATH (Sillitoe et al., 2015), or InterPro (Mitchell et al., 2015) database classification. A general location for the P-CR and CSR domains has been proposed from solution structures of recombinant CatDs of rice CesA8 obtained by small-angle X-ray scattering (SAXS; Olek et al., 2014). The SAXS-derived molecular envelope showed a two-domain structure, with the extended domain participating in dimerization. The best fit of the CesA8 catalytic core domain homology model to this envelope placed the catalytic core in the central portion of the envelope, with the CSR filling the extended domain and the P-CR capping the catalytic core domain distal to the CSR. Ab initio modeling of a cotton (Gossypium hirsutum) CesA1 CatD (Sethaphong et al., 2013; Slabaugh et al., 2014) produced a structurally conserved catalytic core but offers models for the P-CR and CSR domains inconsistent with the SAXS-derived solution structure (Olek et al., 2014).
Here, we report the structural characterization of a rice OsCesA8 recombinant P-CR domain using SAXS and X-ray crystallography. The crystal structure is an uncommon variant of a coiled-coil domain formed by antiparallel α-helices and a large connecting loop, with aromatic residues stacked between the helices in the hydrophobic core. The P-CR crystal structure fits within a cylindrical, elongated molecular envelope for this domain derived by SAXS. In turn, this molecular envelope for the P-CR fits into the previously determined molecular envelope of the entire CatD of the rice CesA8 in a position that is consistent with the volume assigned to the P-CR in previous work (Olek et al., 2014). The combination of the crystallographic structure of the P-CR, the homology model of the CesA8 catalytic core, and two independent SAXS molecular envelopes for the CesA8 P-CR and CatD has produced a more detailed topological model for this portion of the CesA8 structure.
RESULTS
P-CR Solution Structure and Molecular Envelope
The rice CesA8 P-CR domain (residues 399–523) fused with an N-terminal 8× His tag/maltose-binding protein was cloned and expressed in Escherichia coli. Final purification on a size-exclusion chromatography (SEC) column after tobacco etch virus (TEV) cleavage showed a narrow peak for a homogenous protein with the predicted molecular mass of the P-CR monomer (14.8 kD; Supplemental Fig. S1A). In addition, circular dichroism (CD) spectra (195–260 nm) of this P-CR sample indicated a well-folded protein with primarily α-helical secondary structure in solution (Supplemental Fig. S1B).
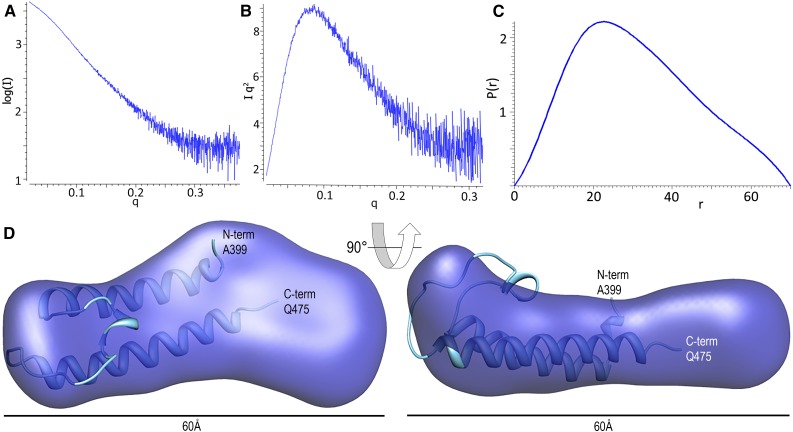
For SAXS studies, an N-terminal 6× His-tagged P-CR domain was expressed and purified in a similar manner, again resulting in a single band with the monomeric molecular mass appropriate for the longer tag in this vector (18.2 kD). Analysis of solution scattering curves corresponding to P-CR monomers gave a radius of gyration (Rg) of 24.2 Å. The Kratky plot of the scattering data showed characteristic peaks and troughs expected for a compact protein structure with a single conformation in solution (Fig. 2, A and B). The molecular envelope of the P-CR was determined from reconstruction calculations using data in the resolution range 0.02 < q < 0.38 Å–1. The final molecular volume (Fig. 2D) was based on averaging dummy residue models obtained from 40 independent reconstruction runs (Supplemental Fig. S2). The averaged P-CR molecular envelope showed an elongated, cylindrical structure with a maximum corner-to-corner distance of 70 Å (Fig. 2C), corresponding to the length of the reconstructed shape of 60 Å (Fig. 2D). The envelope differs significantly on each end of the long axis, with one end displaying a wider, flattened surface while the other shows a more compact structure featuring a distinct protrusion perpendicular to the long axis (Fig. 2D). Combined data from the CD and SAXS experiments indicate that the P-CR possesses an independently folded, compact monomeric structure containing a significant amount of α-helical secondary structure.
Figure 2.
Solution X-ray scattering experiments and determination of the P-CR three-dimensional surface contour structure. A, Log(I) versus q plot of the raw data of the P-CR. B, Kratky plot of the same P-CR data. C, The P(r) pair distribution function for intensity data corresponding to P-CR as calculated by GNOM (Svergun, 1992). D, Three-dimensional molecular envelopes of the P-CR (dark blue) reconstructed from the solution scattering data using the ATSAS software package (Konarev et al., 2006) with GASBOR (Svergun et al., 2001). Molecular surfaces generated from the grid objects that were obtained by averaging multiple reconstruction runs were rendered with Chimera (Pettersen et al., 2004). The P-CR crystal structure (light blue) is shown embedded in its best-fit orientation using MIFit (https://github.com/mifit/mifit).
P-CR Crystal Structure and Architecture
The recombinant P-CR protein, purified as described for the CD experiment, was crystallized in 1 m trisodium citrate/dihydrate, pH 8.6, at 4°C, producing tetragonal bipyramid-shaped P-CR crystals with dimensions of approximately 250 × 100 × 100 μm. Initial local data collection indicated that these crystals had an F23 space group with two P-CR molecules per asymmetric unit (Matthews coefficient of 2.41, 49% solvent content) and unit cell parameters of a = b = c = 150.8 Å (Table I). A survey of lower symmetry space groups confirmed F23 as the optimum choice. Data collected from the selenomethionine (SeMet)-substituted P-CR crystals extended to 2.4 Å and confirmed the crystal packing (Table I). Of the possible eight SeMet residues (four Mets per monomer), only four were observed in an anomalous scattering difference map. Using these four SeMet positions, the structure was solved using single-wavelength anomalous dispersion (SAD) phasing (Dauter et al., 2002). The initial model revealed a 2-fold symmetrical dimer in the asymmetric unit, where 70 of the 125 residues could be readily placed in the initial electron density map.
Table I. Data collection and refinement statistics.
| Parameter | OsCesA8 P-CR (399–523) SeMet (PDB code 5JNP) |
|---|---|
| Data collection | |
| Location | Advanced Photon Source 23-ID-B |
| Energy (Å) | 0.9794 |
| Space group | F23 |
| Cell dimensions | |
| a = b = c (Å) | 150.8 |
| α = β = γ (°) | 90 |
| Resolution (Å) | 45.47–2.40 |
| No. of reflections | 505,517 |
| No. of unique reflections | 21,575 |
| No. of Rfree reflections | 1,118 |
| CC1/2 | 96.3 (79.7) |
| I/σ | 101.4 (2.4) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 45.2 (45.5) |
| Refinement | |
| Total no. of atoms (average B) | 1,366 (86.03) |
| No. of protein atoms (average B) | 1,310 (85.06) |
| No. of waters (average B) | 8 (85.52) |
| No. of ligand/ion atoms (average B) | 48 (113.17) |
| Rwork/Rfree | 0.240/0.272 |
| Root-mean-square deviation for bonds (Å) | 0.009 |
| Root-mean-square deviation for angles (°) | 1.24 |
| MolProbity overall score | 1.16 (100th percentilea) |
| Rotamer outliers | 0% (0) |
| Ramachandran | |
| Preferred | 100% (150) |
| Generous | 0% (0) |
| Disallowed | 0% (0) |
The 100th percentile is the best among structures of comparable resolution; the 0th percentile is the worst.
Given that a significant number of residues in the C-terminal portion of the P-CR domain were missing in the electron density map, mass spectrometry experiments were done on thoroughly washed crystals to determine whether a cleaved P-CR molecule had crystallized. Coverage of the P-CR sequence by detected peptides was 95% and showed that the full-length molecule was in the crystal (Supplemental Fig. S3). SeMet-substituted P-CR crystals also were tested and showed the appropriate shift for the selenium/sulfur substitution for all Mets detected.
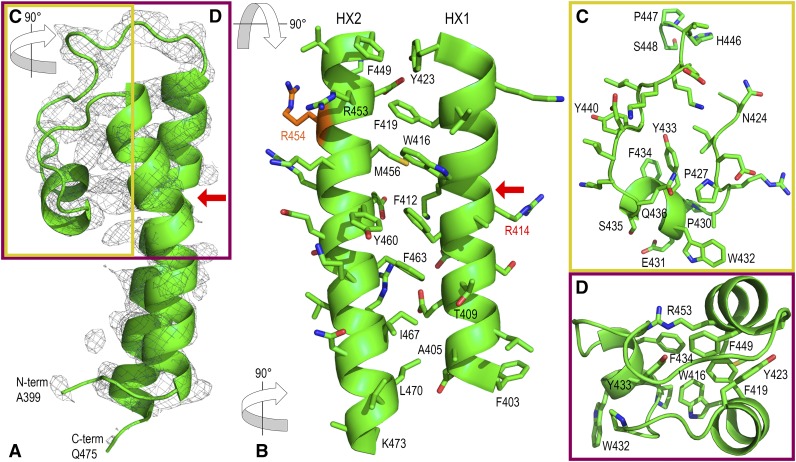
Further refinement of the SAD-phased P-CR structure was done with Rosetta-guided phase improvement (DiMaio et al., 2011; Terwilliger et al., 2012) using a poly-Ala backbone model of the original 70 residues to reduce phase bias in the molecular replacement. This procedure generated an improved electron density map in which the structure could be extended to 77 residues with iterative cycles of building and refinement. Several ligands and water molecules were then placed, and the model was refined to give a final structure for residues 399 to 475 of the P-CR, with C-terminal residues 476 to 523 disordered in the crystal. The structure was evaluated using MolProbity (Chen et al., 2010) and deposited in the PDB with PDB code 5JNP (Table I; Fig. 3A).
Figure 3.
X-ray crystal structure of the rice CesA8 P-CR (Ala-399 to Gln-475). A, Ribbon model of the P-CR crystal structure (green) encased in an electron density omit map (2σ; gray mesh). Colored boxes and rotation arrows depict areas inset in C (yellow) and D (purple), respectively. The red arrow (A and B) points to the bend in HX1 between Arg-414 and Pro-418. B, Details of the interactions between HX1 and HX2, showing R groups and highlighting knob-in-hole interactions. In this orientation, Arg-414 and the stacked Arg-454 and Arg-458 are exterior facing functionalities; Arg-454 (orange) is mutated to Lys in the cellulose-deficient fra6 mutant (Zhong et al., 2003). C, Structure of the P-CR LP region, highlighted in the yellow box in A and rotated 90° relative to A to highlight loop structures. D, Structure of the upper region of the P-CR hydrophobic core, highlighted in the purple box in A and rotated 90° relative to A to highlight the hydrophobic core. Aromatic residues from HX1, HX2, and the LP are tightly packed, forcing Trp-432 to become solvent exposed.
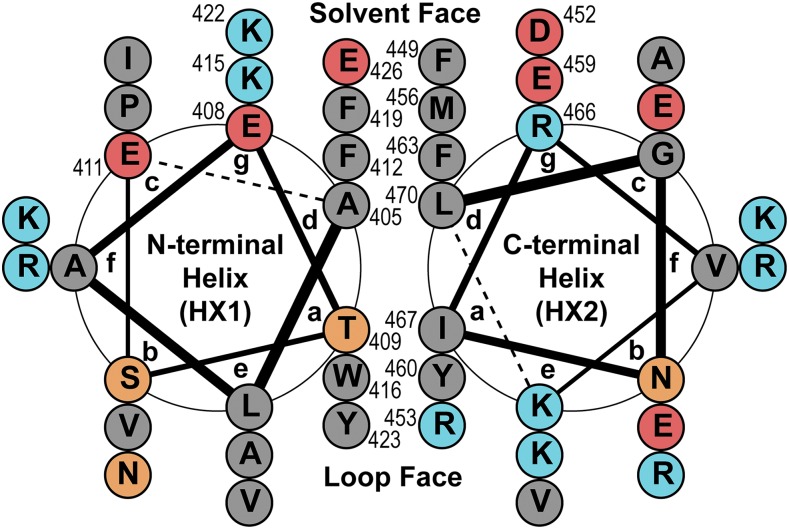
As predicted by CD spectral analysis, the N-terminal region of the P-CR structure is primarily α-helical with a few random coil loops. The structure is dominated by two long antiparallel α-helices that are joined by a 25-residue connector loop that bends back toward the α-helices, forming several hydrophobic contacts (Fig. 3A). The first helix spans approximately 31 Å between residues 403 and 423 (HX1; Fig. 3B), with a bend in HX1 occurring where backbone helix interactions do not occur between Arg-414 and Pro-418 (Fig. 3, A and B). The second large helix (HX2) runs antiparallel to HX1, spanning approximately 35 Å between residues 449 and 473 (Fig. 3B). The hydrophobic core that stabilizes the structure of the P-CR coiled-coil is unusual for its seven stacked aromatic residues, with only three aliphatic residues, a Thr and a Met occupying knob-in-hole positions (Figs. 3B and 4). These aromatic residues force the helices slightly farther apart, relative to standard aliphatic coiled-coils, to accommodate a larger hydrophobic core. The bend in HX1 between Arg-414 and Pro-418 also facilitates interhelical contacts between these larger aromatic residues.
Figure 4.
Helical wheel diagram of the coiled-coil interactions between HX1 and HX2. In an antiparallel coiled-coil, the a and d positions of each helix intercalate to form the hydrophobic core. Compared with most coiled-coils, the P-CR has an uncommonly high proportion of aromatic residues in the a and d positions, which are typically occupied by aliphatic residues. The e and g positions typically form salt bridges between helices, but in the P-CR only the solvent-exposed face appears to form these (Supplemental Fig S4). This figure was illustrated with DrawCoil 1.0 (Grigoryan and Keating, 2008). Circled residue abbreviations are filled with color based on R group properties: hydrophobic (gray), polar neutral (orange), charged acidic (red), and charged basic (blue).
The 25-amino acid (424–448) loop region (LP) connecting the HX1 and HX2 helices is well ordered in the crystal by contacts it makes with HX1 and HX2 (Fig. 3, A and C). A small α-helix within LP between residues 431 and 435 contains a triplet of conserved hydrophobic residues, Trp-432, Tyr-433, and Phe-434. Tyr-433 and Phe-434 face inward toward the α-helices and the aromatic stack of the hydrophobic core between HX1 and HX2 (Fig. 3D). The third conserved residue, Trp-432, is fully solvent exposed and shows only weak electron density (Fig. 3, C and D).
Although knob-in-hole interactions form the basis of coiled-coil structures, electrostatic interactions also can determine the orientation of helices within the coiled-coil or restrict interactions to specific binding partners (Grigoryan and Keating, 2008). The antiparallel helices of the P-CR include three regions with potential electrostatic interactions along the length of the coiled-coil, although most of these interactions are approximately 4 Å and longer than a typical H-bonding distance (Supplemental Fig. S4). All of these amino acids are located on the solvent face of the coiled-coil and opposite the side that interacts with LP.
The DALI (Holm and Rosenström, 2010), 3D-surfer (La et al., 2009), and BioX.GEM 3D-Blast (Tung et al., 2007) algorithms all showed consistent alignment of the P-CR with other coiled-coil proteins. The closest matches were the pore-forming domains of several voltage-gated sodium channel structures (Catterall, 2014). These structures also have numerous aromatic residues stacked between the helices and a large connector loop that occupies approximately the same position as does the P-CR. However, because the biological role of helices lining sodium channels appears distinct from the possible roles of the helices in the P-CR, this similarity is not likely to extend beyond the structural level.
Comparison of the P-CR Crystallographic Structure with the SAXS Data
The partial X-ray crystallographic structure of the P-CR was fit into the SAXS molecular envelope of the protein in solution. The best-fit orientation of the coiled-coil of the P-CR crystal structure into the SAXS molecular envelope left a substantial density that would contain the 48 unobserved C-terminal residues and any flexible region of the N-terminal tags (Fig. 2D). The SAXS-based reconstruction methodology involves the imposition of relatively flat protein density that is reliable at low resolution (approximately 20 Å). In contrast to atomic X-ray crystallographic data, the reconstructed molecular shapes are relatively insensitive to atomic movements on a scale that would render atoms invisible in the 2.4 Å resolution electron density map. For example, a SAXS-based envelope should resolve atomic groups fluctuating with displacements of approximately 5 Å, but atoms associated with structural variations of this magnitude would be invisible in a crystallographic map. As noted earlier, the Kratky plot (Fig. 2B) shows a relatively large maximum at q of approximately 0.08 Å–1 and a small rise at higher q, suggesting that the P-CR structure was well folded under the conditions used for the solution scattering experiments. Furthermore, calculations of scattering patterns with CRYSOL (Svergun et al., 1995) from the crystal structures of both the monomer and the pair of molecules in the crystallographic unit gave poor fits to the scattering data, and the resulting values of Rg were much smaller than those indicated by the direct analysis of SAXS data (Rg = 24.2 Å according to data analysis with GNOM [Svergun, 1992] but χ = 29.1, Rg = 14.6 Å and χ = 32.3, Rg = 18.3 Å for the crystal monomer and dimer models, respectively). Because the crystal structure contains only two-thirds of the P-CR sequence, lack of agreement would be expected as the SAXS scattering data corresponds to the complete structure rather than the partial model. In addition, a disorder propensity calculation for the P-CR sequence using the GLOBPLOT2 server (Linding et al., 2003; http://globplot.embl.de) predicted that the N-terminal sequence is more ordered than the C-terminal sequence and shows a sharp disorder maximum at the point in the sequence where the electron density in the crystallographic map disappeared. This predicted flexibility suggests that the C-terminal domain might occupy multiple conformational states within the confines of the crystal lattice, rendering it invisible in the atomic resolution electron density map. Therefore, the isolated P-CR molecule appears to contain two loosely linked structural domains with a C-terminal domain that is intrinsically less well ordered than the N-terminal domain.
Molecular Docking of the P-CR with the Catalytic Core
By combining the crystallographic structure of the P-CR sequence with the SAXS-derived molecular envelope of the entire P-CR volume, a homology-based chain trace of the catalytic core, and our previous reconstruction of the CesA8 CatD molecular envelope (Olek et al., 2014), we developed a detailed topological model of the CesA8 structure. The P-CR model was aligned with the SAXS-based P-CR reconstruction, representing the partial specific volume of the entire P-CR domain. The most plausible fit placed the P-CR loop region into the end of the volume with the distinct protrusion perpendicular to the main axis of the envelope, which then encompassed the helical coiled-coil (Fig. 2D). Unoccupied envelope regions presumably include the C-terminal residues that are disordered in the crystal (Lys-476 to Val-523). Using this best fit and the crystal structure of the P-CR as a guide, the molecular SAXS envelope was placed into the F23 crystal structure, resulting in good packing with no significant molecular overlaps (Supplemental Fig. S5).
To place the P-CR in the context of the entire CatD, the crystallographic P-CR structure within its SAXS molecular envelope was fused with the CesA8 catalytic core homology model (lacking the P-CR and CSR), and both objects were fit to the SAXS-derived molecular envelope of CesA8 CatD obtained in previous work. In this earlier interpretation, only three plausible orientations of the CesA8 catalytic core homology model were identified, all of which occupied the central region of the CesA8 CatD molecular envelope, as this is the only portion of the envelope wide enough to contain the catalytic core (Olek et al., 2014). These placements leave two disparate volumes on either side of the catalytic core that must be occupied by the P-CR and CSR domains. CesA8 sequence continuity required that the N terminus of the P-CR connect directly to the catalytic core two residues after the first catalytic residue (Asp-396) that binds the uracil in the UDP-Glc substrate, a relatively fixed point in the structure (Morgan et al., 2013). Constraining the N terminus of the P-CR model to this end point eliminated most translational degrees of freedom in the placement of the P-CR relative to the catalytic core and determined that the P-CR domain occupied the density as shown in Figure 5A. Fitting of the P-CR molecular volume into the CesA8 CatD volume was achieved by matching the length of the P-CR SAXS reconstruction to the length of the larger CesA8 CatD (Fig. 5B). The unoccupied extended domain of the CatD molecular envelope was then consistent with the position on the catalytic core where the CSR domain would be inserted. In this new topological model, the catalytic core model was not frozen at the position determined in previous work but was refit to incorporate the new constraints and structural information from the P-CR data.
Figure 5.
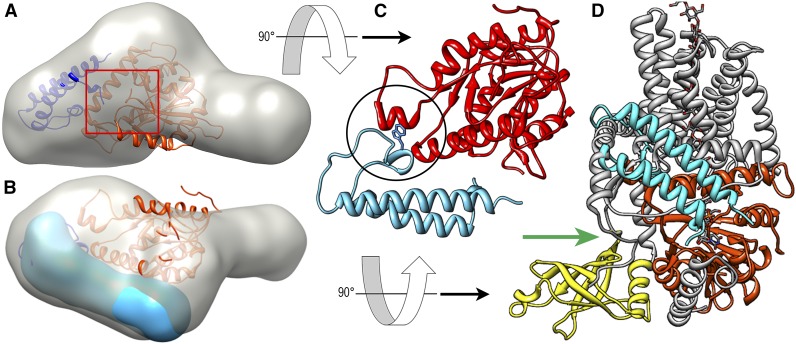
Best-fit models of the P-CR in the OsCesA8 CatD. A, The P-CR fit in the three-dimensional molecular envelope of the CatD (gray; Olek et al., 2014) using MIFit (https://github.com/mifit/mifit). The CatD homology model (orange) and P-CR crystal structure (navy blue) are shown in their best fit, which places the N terminus of the P-CR near the junction with the catalytic core (red square). B, Rotated view shows the three-dimensional molecular envelope of the P-CR (blue) overlaid in the best-fit orientation. C, A potential hydrophobic contact with Trp-432 (stick) between the catalytic core and the P-CR LP region (black circle), illustrated here with P-CR (light blue) and the OsCesA8 CatD homology model (red). D, The CatD homology model alignment with the BcsA P-CR coiled-coil (light blue) is overlaid onto the BcsA crystal structure. CesA homologous regions (orange) and nonhomologous regions (gray) place the termini near the substrate entry pathway of BcsA (green arrow; Morgan et al., 2013, 2014). The position of the BcsA PilZ domain (yellow) does not overlap that of the P-CR, which is found on the opposite side of the catalytic core domain.
The modeling of the P-CR domain with the CesA8 catalytic core placed some of the P-CR LP residues in a hydrophobic pocket in the catalytic core, eliminating the solvent exposure of Trp-432 (Fig. 5C). As a result, the hydrophilic residues on the opposite face of the coiled-coil were placed in a solvent-exposed position on the exterior of the CesA8 catalytic core. Several of the hydrophilic residues of the P-CR near the junctions of HX1 and HX2 with the LP region, the most distal point from the start of the P-CR domain insertion, were near the predicted membrane bilayer based on the position of the transmembrane α-helices in the homologous BcsA structure (Fig. 5D). However, the best fit did not indicate any hydrophobic membrane or transmembrane associations.
In an independent trial, attempts were made to fit the P-CR structure into the small extended domain of the CesA8 CatD molecular envelope, which we previously predicted to contain the CSR domain (Olek et al., 2014). Attempts to fit the P-CR structure into this extended domain resulted in severe clashes between the P-CR volume and the model of the catalytic core domain, demonstrating that this volume is too small to accommodate the P-CR (Supplemental Fig. S6). Any movement of the catalytic core domain required to allow the P-CR to occupy the small domain, while eliminating the overlaps with the catalytic core, pushes the catalytic core and CSR domains substantially out of the molecular envelope. For this reason, this placement of the P-CR domain appears inconsistent with the SAXS data and confirms the placement in the position shown in Figure 5.
DISCUSSION
The P-CR Domain Is a Unique Structure in Plant Cellulose Synthases
Solving the crystal structure of the P-CR and docking its location within the rice CesA8 CatD represents a major step in understanding the assembly of a CSC. The P-CR domain of CesA is a conserved feature of plant cellulose synthases not found in their bacterial homologs. Structural characterization of the P-CR shows that this domain comprises an antiparallel coiled-coil that incorporates an uncommonly high proportion of aromatic residues into a knob-in-hole stabilizing structure. This aromatic stack creates an expanded hydrophobic core between helices that forms several contacts with a large connecting loop, a rare feature for coiled-coils. The CesA8 catalytic core is a strongly conserved, novel variant of a classic nucleotide-binding fold found in several types of processive glycosyl transferases (Olek et al., 2014). Modeling based on sequence and structural data shows that the P-CR locks into this catalytic core, aligning the coiled-coil domain along the surface, with one end at the entrance to the active site of the CatD and the other near the predicted membrane-spanning domains (Fig. 5).
Data-Driven Modeling of the CesA8 CatD
Previous atomic models of the CesA CatD have been based solely on ab initio computational modeling of the core, P-CR, and CSR domains (Sethaphong et al., 2013). We have derived a three-dimensional model of the CesA8 CatD by combining experimental structural information from SAXS-derived solution structures of the recombinant CesA8 CatD and P-CR and the crystal structure of the P-CR. Explicit assumptions for this approach were (1) that the CesA8 and BcsA catalytic cores are structurally similar and (2) that the P-CR domain adopts the same fold in isolation as in the full-length structure. Regarding assumption 1, the catalytic core domains of bacterial type II synthases homologous to plant CesAs are remarkably similar in structure based on root-mean-square deviation and TM scores (Olek et al., 2014). In addition, key sequence motifs that are critical for substrate binding and catalysis, as well as those that interact with the developing polysaccharide chain, are common between BcsA and CesAs. Although the exact shape of the catalytic core of CesA might differ slightly from that of BcsA, the elements integral to synthesizing the polysaccharide chain are unlikely to vary substantially between these structures. While we cannot completely rule out the possibility that the P-CR structure is refolded in the context of the CesA structure, the robustness of Kratky plots of the solution structure of the P-CR (Fig. 2) and the fit of the crystal structure into this component are indicative of a well-folded P-CR structure. It seems unlikely that such an energetically favorable and stable structure would adopt a different conformation in the CesA8 CatD.
Plausible arrangements for the P-CR and catalytic core components within the CesA CatD envelope are restricted by the dimensions of the SAXS-based envelope for the P-CR (Figs. 2 and 5). Our P-CR reconstruction resulted in an elongated molecular shape of the same length as the long dimension of the larger CesA8 CatD structure obtained in our earlier SAXS-based reconstruction (Olek et al., 2014). We showed previously that the bulky catalytic core must occupy a specific orientation within the central region of the CesA envelope (Olek et al., 2014). This placement precludes positioning the P-CR within the smaller domain because the elongated molecular envelope would then penetrate into the space occupied by the catalytic core (Supplemental Fig. S6). The only non-occluded space in which the P-CR envelope can be accommodated lies against one edge of the larger domain in the CesA envelope (Fig. 5). Thus, the shapes and dimensions of the catalytic core and P-CR are sufficient to map out their respective spaces within the larger domain of the CesA CatD envelope. The unfilled space of the smaller domain is then of an appropriate size to contain the CSR. In addition, we demonstrated previously that dimerization is mediated by interaction surfaces on the smaller domain (Olek et al., 2014); we have also demonstrated that the P-CR shows no propensity to dimerize (Supplemental Fig. S1A).
An atomic-level model for the larger CesA domain is obtained via consideration of the fit of the P-CR crystal structure into the SAXS-derived P-CR molecular envelope. Placement of the P-CR partial crystal structure at the end of the SAXS-based P-CR envelope is dictated by the shape of the envelope (Fig. 2). A systematic evaluation determined the most plausible fits of the catalytic core into the CesA envelope with the requirement that the chain trace is almost completely contained within the CesA envelope (Olek et al., 2014). However, only one orientation of the catalytic core and of the P-CR envelope/model within the CesA8 envelope allows the required sequence connectivity between these two components (Fig. 5). Since the point of connection of the P-CR sequence to the CesA sequence lies immediately adjacent to the first key sequence motif (SDDG), we consider this attachment point fixed relative to the catalytic core. In fact, it would be difficult to justify a model that required a shift in the section of the polypeptide incorporating this motif, because that change would eliminate a critical interaction with the UDP substrate.
Although the level of detail incorporated into this assembled model is limited by the relatively low resolution of the SAXS-based envelopes and potential differences between the BcsA and CesA catalytic cores, the model incorporates all available information in the most direct and plausible way. The end product is a topological model of the CesA8 CatD that is sufficiently detailed to make testable hypotheses using site-directed mutagenesis to probe potential interaction surfaces.
Potential Functions of the P-CR Domain
Of the two plant-specific sequences in the CatD, only the P-CR has not previously been assigned a possible functional role in vivo. Because the bacterial synthase lacks the P-CR and CSR, these plant-specific sequences are unlikely to function directly in catalysis. The N-terminal helix of the P-CR coiled-coil connects with the catalytic core two residues after the first nucleotide-binding catalytic motif at Asp-396 near the substrate entry site, but the coiled-coil extends well away from this catalytic amino acid and is docked into hydrophobic contacts distant from the active site. However, docking of the coiled-coil domain of the P-CR at the entrance to the active site might suggest a role in the control of substrate delivery in the absence of the bacterial PilZ domain (Morgan et al., 2013). In BcsA synthase, cyclic di-GMP activation of the PilZ domain interacts with a gating loop to uncover the entrance to the active site (Morgan et al., 2014). An FxVTxK motif in the gating loop coordinates the UDP moiety of the nucleotide sugar in the bound state (Morgan et al., 2016). The FxVTxK motif is conserved in all plant CesAs, but traditional prediction of eight transmembrane helices (TMH) would place the motif in a large extracellular loop between TMH5 and TMH6. In a more recent evaluation of predictions of transmembrane domain structure, TMH5 is proposed to occupy an interfacial position, placing the FxVTxK motif in a cytosolic location where it could function similarly to the bacterial enzyme (Slabaugh et al., 2016). Substitution of the Val and Lys residues of the motif results in severe disruption of cellulose synthesis (Slabaugh et al., 2016). Recent work employing limited proteolysis of tagged CesAs confirms that the FxVTxK motif is located in the cytosol and is not extracellular (C. Haigler, personal communication). Given this new placement of TMH5, an interaction with the P-CR might mediate substrate delivery.
The P-CR also could play a role in the assembly of the CSC by mediating contacts with CatDs of neighboring CesAs in the complex. Recombinant soluble rice CesA8 CatDs are thought to dimerize through their CSR domains (Olek et al., 2014). As described earlier, dimers of P-CR are observed in the crystal, but their relative orientation is problematic in the context of the full CatD, which would force the P-CR loop region to clash significantly with the other monomer’s catalytic core (Supplemental Fig. S7). More recently, recombinant Arabidopsis (Arabidopsis thaliana) CesA1 CatDs renatured in detergent from inclusion bodies were shown by SAXS modeling to form trimeric structures (Vandavasi et al., 2016). Attempts to fit ab initio models based on the cotton CesA1 CatD (Sethaphong et al., 2013) using either P-CR or CSR symmetrical contacts exhibited displacements of the CesA model out of the molecular envelope (Vandavasi et al., 2016). However, the SAXS molecular envelope of native recombinant monomeric CesA8 CatDs determined by Olek et al. (2014) was assembled into a trimer with the appearance and dimensions that resemble the trimeric assembly depicted by Vandavasi et al. (2016; Supplemental Fig. S8A). In this interpretation, the extended CSR domains fill the corners of the trimer and the P-CR domains form the contacts between the CatDs, consistent with the model of Vandavasi et al. (2016) but contrary to CSR trimeric contacts proposed by Sethaphong et al. (2016). The P-CR contacts in this model conform to a 3-fold contact in the crystal structure formed between pairs of Arg residues (Arg-454 and Arg-458) that are mediated by a phosphate ion (Supplemental Fig. S8B). However, the trimeric model stabilized by P-CR domains is likely to be possible only when the membrane-spanning domains are deleted from the recombinant protein. Expanding the P-CR trimer structure using the model of a single P-CR docked to the CesA8 CatD results in a severe clash with the currently proposed position of the membrane domains (Supplemental Fig. S8C). Thus, while a tight interaction of P-CR domains in isolated CatDs explains the formation of homomeric trimers in vitro, the clashes that would result with the membrane-spanning domains in the full-length protein bring into question any biological relevance in vivo.
Another possibility for the role of the P-CR is in binding to accessory proteins. The membrane-associated endo-(1→4)-β-d-glucanase KORRIGAN required for proper cellulose synthesis is one such candidate, but little information is known about the function or location of this protein (Mansoori et al., 2014). Microtubule-binding proteins that interact with CesA, cellulose synthase interactive proteins (CSI1 and CSI3; Li et al., 2012; Lei et al., 2013), are additional candidates, but the location of the P-CR at the substrate entry portal to the active site would seem to make these interactions less likely. A location at the entrance to the active site also might suggest an association with proteins such as sucrose synthase, which has long been proposed to form metabolic channels to deliver UDP-Glc into the active site (Amor et al., 1995).
Arg-454 participates in an unusual stacked trimeric contact in the crystals, stabilized by a phosphate ion (Supplemental Fig. S8B). This residue is interesting, as a nonsynonymous fra6 mutation in Arabidopsis converts Arg-454 to Lys, resulting in the disruption of normal cellulose synthesis (Zhong et al., 2003). However, Arg-454, which participates in the crystallographic trimer in vitro, is oriented toward the predicted hydrophilic exterior of the HX1-HX2 coiled-coil (Fig. 3B), where interaction with other synthase complex components also is possible. This finding underscores the potential importance of this P-CR residue in synthase function but does not yield specific information on the types of interactions in which it may participate.
Uncertainty Remains about the Number of CesAs per CSC
The number of (1→4)-β-d-glucan chains in a cellulose microfibril, and so the number of CesA catalytic units in a synthase complex, remains undetermined. Classical measurements of microfibril diameters between 3 and 4 nm indicated that 36 chains were assembled by the hexameric rosette complexes, indicating a hexamer of CesA hexamers. More recent solid-state NMR measurements of cellulose crystalline cores and surface chains suggest that cellulose microfibril diameters range from 2.3 to 3 nm, corresponding to 18 to 24 glucan chains (Fernandes et al., 2011; Newman et al., 2013; Thomas et al., 2013) and potentially a smaller CesA oligomer. Genetic functional analyses of CesA mutants established that three distinct isoforms of CesAs are required for optimal cellulose synthesis and are developmentally regulated to be specific for primary cell wall and secondary cell wall synthesis (Taylor et al., 2004; Taylor, 2008). Quantitative antibody-labeling studies indicated that these isoforms are present in 1:1:1 ratios (Gonneau et al., 2014; Hill et al., 2014), prompting these authors to propose that a rosette comprises six specific heterotrimer interactions that account for the stoichiometry and for the synthesis of an 18-chain microfibril. Consistent with this interpretation, image-enhancement techniques applied to the external portions of the transmembrane domains of CesA rosettes observed in freeze-fracture transmission electron microscopy give high proportions of triangular particle counts in these hexameric structures (Nixon et al., 2016). Vandavasi et al. (2016) argued that two homotrimers of each of the three isoforms also preserve the stoichiometry. However, neither model explains how microfibrils larger than 18 chains can be made. On the other hand, there is a wealth of data supporting the formation of both homodimers and heterodimers in vivo (Taylor et al., 2003; Desprez et al., 2007; Timmers et al., 2009) and in even-numbered monomeric units (Atanassov et al., 2009), which provides many options for preserving this isoform stoichiometry in homodimer and heterodimer combinations. While the homotrimer formation of the isolated CatDs appears to involve coupling of the P-CR, specific models of this interaction are unlikely to represent a biologically relevant interaction, given the structural clashes predicted when the CatD is placed in the context of the predicted location of the membrane-spanning domains (Supplemental Fig. S8C). The structure of the P-CR and its predicted location in the CatD have given us new insight into the formation of the CSC, but we need an atomic resolution structure of an entire CesA alone or in an intact rosette in order to resolve these questions at the core of understanding cellulose synthesis in plants.
MATERIALS AND METHODS
Cloning and Heterologous Protein Expression
For SAXS analysis, a pBluescript SKII vector (Stratagene) containing the rice (Oryza sativa) OsCesA8 cDNA (LOC_Os07g10770; formerly called OsCesA13) was used as a template to clone the P-CR, Ala-399 to Val-523. The 375-bp P-CR DNA insert was amplified by PCR using a forward primer (5′-GCGCCATGACTAGTGACTCGGCAATGCTGACTT-3′) and a reverse primer (5′-GTGGTGCTCGAGTTACTAGACCAGGCGGGGAA-3′) ordered from Integrated DNA Technologies. The P-CR was ligated into a modified pETM-11 (EMBL) vector, containing an N-terminal 6× His tag, using SpeI and XhoI restriction sites. The empty pETM-11 plasmid was obtained from EMBL (http://www.embl-heidelberg.de). The OsCesA8 P-CR pETM-11 plasmid construct was transformed into competent Escherichia coli [Rosetta2 (DE3); Novagen] cells using heat shock. Positive transformants were selected with 50 μg mL–1 kanamycin and 25 μg mL–1 chloramphenicol and then used to inoculate 5 mL of Luria-Bertani medium for overnight growth. The 5-mL overnight culture was used to inoculate a 500-mL Luria-Bertani culture that was grown to 0.6 to 0.8 optical density at 600 nm and then cooled to 16°C before recombinant protein induction with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 13 h.
For CD and crystallization experiments, a higher yield construct was desired, so a vector that includes a cleavable maltose-binding protein and a His tag was used to produce a recombinant CesA8 P-CR domain. The rice OsCesA8 full-length cDNA sequence template was synthesized with E. coli-optimized codons and cloned into the pUC-57 vector using the SmaI restriction site (Bio Basic). The OsCesA8 P-CR, Ala-399 to Val-523, was amplified by PCR using a forward primer (5′-TACTTCCAATCCAATGCCGCTATGCTGACCTTCG-3′) and a reverse primer (5′-TTATCCACTTCCAATGCTATCATTAGACCAGACGAGGC-3′) ordered from Integrated DNA Technologies. This was cloned into a pEV-L9 vector (E. Harms and R. Kuhn, unpublished data) containing an N-terminal 8× His tag, a maltose-binding protein fusion, and a TEV protease site using ligation-independent cloning (Aslanidis and de Jong, 1990). The OsCesA8 P-CR pEV-L9 plasmid construct was transformed into competent E. coli (DE3) cells using heat shock. Positive transformants were selected with 50 μg mL–1 kanamycin and then used to inoculate 10 mL of Luria-Bertani medium for overnight growth. The 10-mL overnight culture was used to inoculate a 2-L Luria-Bertani culture that was grown to 0.4 to 0.6 optical density at 600 nm and then cooled to 20°C before recombinant protein induction with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 18 h.
For crystal phasing seleno-l-Met-substituted (Ramakrishnan and Biou, 1997) P-CR protein was obtained using the same OsCesA8 P-CR pEV-L9 transformed cells and expression conditions, with the substitution for Luria-Bertani broth of a medium containing M9 salts, pH 7.4, 0.4% Glc, 2 mm MgSO4, 25 μg mL–1 FeSO4, 1 μg mL–1 vitamins (riboflavin, niacinamide, pyridoxine monohydrochloride, and thiamine), 40 μg mL–1 basic l-amino acids, except Met, and 40 μg mL–1 seleno-l-Met.
Protein Purification
For SAXS experiments, frozen cell pellets containing the induced OsCesA8 P-CR pETM-11 construct were suspended in buffer solution containing 50 mm HEPES[HCl], pH 8, 500 mm NaCl, 20 mm imidazole, 0.2% Nonidet P-40 detergent, and 0.5× cOmplete, Mini, EDTA-free protease inhibitor cocktail (Roche) and then lysed by sonication. Cell debris was removed by centrifugation, and the soluble protein supernatant was applied to Ni2+ 6-Fast Flow resin microbeads (GE Healthcare) and incubated for 1 h, pelleted, and washed with 50 mm HEPES[HCl], pH 8, containing 1 m NaCl and 20 mm imidazole. Protein was eluted from the microbeads by incubating the beads in an elution buffer solution containing 50 mm HEPES[HCl], pH 8, containing 0.5 m NaCl and 300 mm imidazole. Eluted protein was injected onto a Superdex 200, 10/300GL (GE Healthcare) SEC column equilibrated in 50 mm HEPES[HCl], pH 8, containing 0.5 m NaCl, to further purify the sample, producing a monomeric, homogenous sample for SAXS analysis.
For CD and crystallization experiments, frozen cell pellets containing the induced OsCesA8 P-CR pEV-L9 construct were suspended in base buffer (50 mm HEPES[HCl], pH 8, and 0.2 m NaCl) containing 20 mm imidazole and 1× Halt Protease Inhibitor Single-Use Cocktail, EDTA-free (100×; Thermo Fisher Scientific) and then lysed using a French pressure cell. Cell debris was removed by centrifugation, and the soluble protein supernatant was injected onto a 5-mL HiTrap Chelating HP column (GE Healthcare) equilibrated at 4°C in base buffer containing 20 mm imidazole (binding buffer). The column was washed with binding buffer, and the bound protein was eluted with a linear gradient of 20 to 500 mm imidazole in base buffer. Fractions containing the OsCesA8 P-CR pEV-L9 protein were combined and dialyzed overnight at 4°C in binding buffer to remove excess imidazole while simultaneously digesting the OsCesA8 P-CR pEV-L9 protein with TEV protease (1:50 mg of TEV protease:mg of purified protein). Dialyzed, TEV-cleaved protein was reapplied to the HiTrap Chelating HP column (GE Healthcare) and equilibrated in binding buffer, and the flow through was collected to isolate the untagged P-CR. P-CR samples were further purified by injection onto a HiLoad 16/600 Superdex 200 pg (GE Healthcare) SEC column equilibrated at 4°C in base buffer.
CD Analysis of P-CR Secondary Structure
Pure, TEV-digested P-CR was concentrated to 1.5 mg mL–1 in base buffer using Ultracel-10K Amicon Ultra Centrifugal Filters (Millipore). P-CR samples were loaded into a quartz spectrophotometer cell with a 0.1-mm path length (Starna Cells), and spectra were collected on a Chirascan CD Spectrophotometer (Applied Photophysics) at 22°C. P-CR spectra were collected by scanning between 195 and 260 nm, and the millidegree output was converted to mean residue ellipticity. The P-CR mean residue ellipticity spectra was used to evaluate the proportion of secondary structure with the K2D deconvolution algorithm (Andrade et al., 1993) on the DichroWeb server (Whitmore and Wallace, 2004).
SAXS Data Collection and Processing
All data were collected at the BioCAT undulator beamline (18ID) at the Advanced Photon Source with a 3.5-m SAXS camera as described previously (Fischetti et al., 2004). Data were collected using a sample cell consisting of a thin-walled quartz capillary held at an ambient temperature of 4°C. To minimize radiation damage, protein samples were made to flow through the X-ray beam at a rate that limited X-ray exposure of any one protein to approximately 100 ms. At these exposure levels, the effect of radiation damage on radiosensitive test proteins is undetectable. Typically, a data set consisted of a series of three to five 2-s exposures from protein collected at the peak observed in samples separated by SEC as described above. The estimation of buffer scattering was derived by exposure of the flow through of the column. The two-dimensional scattering patterns were circularly averaged using BioCAT software, and the resulting one-dimensional intensity distribution was plotted as a function of spacing 1/d. sd values of the observed data were calculated, with error propagation formulae used to calculate their effect on the final estimate of scattering from protein. Scattering patterns, including the generation of log(I) versus q plots, Kratky plots, and Guinier plots, were visualized using display programs from the ATSAS software suite (Konarev et al., 2006). The Guinier plot appears approximately linear over the low-q portion of the data used in this study, with Rg of approximately 22.7 Å when estimated using the AutoRg function from the ATSAS data analysis software.
Reconstruction of Molecular Envelopes
The molecular envelope of the P-CR domain was reconstructed using programs from the ATSAS software suite (Konarev et al., 2006). The particle distance distribution function was calculated with GNOM (Svergun, 1992) using a maximum interatomic distance of 70 Å and data in the range 0.02 < q < 0.38 Å–1. Dummy residue models were generated to fit these data using the program GASBOR (Svergun et al., 2001) with the number of dummy residues set equal to the true number of amino acids in the P-CR construct. Forty independent models were aligned for averaging using the SUPCOMB program (Kozin and Svergun, 2001). The 40 aligned models were then averaged to provide a three-dimensional reconstruction of the shape of the P-CR with a partial specific volume set to the expected value from the P-CR sequence. Subaverages of these runs were calculated in order to check the convergence and reproducibility of the final average (Supplemental Fig. S2). Results were visualized using the graphics programs MIFit (https://github.com/mifit/mifit) and Chimera (Pettersen et al., 2004).
Fitting of Crystal Structures to Solution-Scattering Reconstructions
The partial crystal structure of the P-CR was fit to the pseudo-atom grid object representing the P-CR volume using the interactive molecular graphics program MIFit (https://github.com/mifit/mifit). Fitting was guided by the significant difference in the shape at the two ends of the cylindrical reconstruction, where one end is significantly larger and better accommodates the loop that connects the helical regions of the model. To develop a data-driven topology model of CesA, the P-CR SAXS molecular envelope containing the P-CR atomic model was combined with the CatD SAXS molecular envelope and the homology model for the CesA8 conserved catalytic core using MIFit, as described in previous work (Olek et al., 2014). Fitting constraints included matching the shapes of the reconstruction volumes and sequence continuity between the atomic coordinates for the P-CR structure and the homology model of the conserved catalytic core (described above). In this new topological model, the catalytic core model was refit to incorporate the explicit and implicit information from the P-CR and was not frozen at the position depicted by Olek et al. (2014).
Crystallization of P-CR
Pure, TEV-digested P-CR was concentrated in base buffer to a range of protein concentrations, from 5 to 20 mg mL–1 for initial crystallization screening and then 7 mg mL–1 for optimized crystals, using Ultracel-10K Amicon Ultra Centrifugal Filters (Millipore). Crystallization conditions were initially determined with 96-well sitting drop experiments with Classics Suite and JCSG Core Suite crystal screens (Qiagen). Experiments were conducted using a 1:1 protein:reservoir ratio and quickly combined, sealed, and incubated at 4°C with a duplicate at 22°C. After 3 d, crystals were observed only in drops incubated at 4°C containing 0.06 m Na2HPO4, 1.34 m K2HPO4, or 0.96 m trisodium citrate/dihydrate, pH 7, reservoir solution. Crystallization conditions were optimized to increase crystal size and diffraction quality, resulting in optimal crystal growth on a 24 Cryschem M plate (Hampton Research) sitting drop experiment where 1 μL of P-CR protein was combined with 1 μL of 1 m trisodium citrate/dihydrate, pH 8.6, and then incubated for 1 week at 4°C. Cryoprotection was optimized for P-CR crystals, resulting in crystals being slowly introduced to buffer containing 1 m trisodium citrate/dihydrate, pH 8.6, with incremental increases in glycerol concentration up to 20% (w/v). Cryoprotected crystals were then flash frozen in liquid nitrogen, and the diffraction quality was evaluated at the Purdue University Macromolecular Crystallography Facility on a Rigaku-RU200 rotating anode generator with an R-Axis IV++ detector. P-CR crystals with the highest diffraction quality were stored in liquid nitrogen for later data collection at the Advanced Photon Source synchrotron source.
To ensure that crystals contained intact P-CR protein, crystals were dissolved in 2× SDS-PAGE buffer and run on 15% SDS-PAGE gels. The single band corresponding to the P-CR molecular mass (14.8 kD) was excised for each sample and submitted to the Purdue proteomics facility for mass spectrometry fingerprinting.
X-Ray Data Collection, Processing, and Three-Dimensional Atomic Modeling
Data were collected on the high-diffraction-quality, cryoprotected P-CR crystals at the Advanced Photon Source on beamline 23-ID-B. Data were collected in 0.5° oscillation steps over 360° with X-rays at 12.659 keV energy (0.98 Å) using a Mar 300 CCD detector with a 250-mm detector distance. Frames were indexed in the F23 space group, then integrated and scaled to a cutoff of I/σ > 2 using HKL2000 software (Otwinowski and Minor, 1997). SAD (Dauter et al., 2002) frames of the SeMet derivative P-CR crystals were collected at a peak energy of 12.659 keV (0.98 Å). Phenix software (Adams et al., 2010) was used to phase the SAD frames via the AutoSol module (Terwilliger et al., 2012), which placed four SeMet residues in the asymmetric unit and auto fit two P-CR monomer sequences. The two structures were iteratively refined to initial structures containing a chain with 70 of the 125 total residues, with the remaining C-terminal residues unbuilt.
The initial P-CR structure based on SAD phases was iteratively rebuilt with Coot (Emsley and Cowtan, 2004) and refined in Phenix (Adams et al., 2010) to improve geometry and extend the C-terminus. At this resolution, the two P-CR monomers in the asymmetric unit appear identical, so NCS restraints were applied. The results showed modest improvement over independent refinement of the two molecules but did not sufficiently improve the models to allow extension of the C-terminus. Attempts were made to resolve the C-terminal electron density, lacking in the initial structure, by first processing and solving the structure in all lower symmetry space groups, none of which provided any improvement. Using the F23 space group data, a poly-Ala model was extended by three residues into weaker density in the 2Fo-Fc map (2σ) but was unsuccessful, as the subsequent omit maps indicated no significant density improvements. Molecular replacement guided by Rosetta software (Rohl et al., 2004) through the MR-Rosetta module (DiMaio et al., 2011; Terwilliger et al., 2012) in Phenix (Adams et al., 2010) was then tried using a poly-Ala backbone model of the original 70 residues to reduce phase bias in the molecular replacement. Iterative rebuilds and refinement of the MR-Rosetta model in Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010) generated the best map but still lacked complete C-terminal electron density. The improved map allowed for a chain with 77 of the 125 total residues to be built, but the remaining C-terminal residues were not visible and are likely disordered in the crystal. At this point, additional density could be assigned to waters and citrate molecules, and a phosphate molecule and a glycerol molecule were found coordinated on opposite sides of the 3-fold crystallographic axis.
PDB Deposition of the P-CR Structure
The P-CR crystal structure data and coordinates were deposited in the PDB (http://www.rcsb.org/pdb/home/home.do) under PDB code 5JNP.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. P-CR secondary structure and apparent Mr analysis.
Supplemental Figure S2. Reconstructing the P-CR SAXS molecular envelope.
Supplemental Figure S3. Sequence coverage for mass spectrometry data of OsCesA8 crystals.
Supplemental Figure S4. Electrostatic interactions between coiled-coil α-helices HX1 and HX2.
Supplemental Figure S5. The P-CR SAXS molecular envelope fit into the P-CR crystal lattice.
Supplemental Figure S6. Alternative fit of the P-CR into the CesA8 CatD molecular envelope.
Supplemental Figure S7. Noncrystallographic dimer of the P-CR structure.
Supplemental Figure S8. Constructing a P-CR trimer from SAXS and crystallographic data.
Supplementary Material
Acknowledgments
We thank Tim Schmidt, Chun-Liang Chen, and Stanislav Zakharov (Purdue University) for their assistance in this work.
Glossary
- CSC
cellulose synthase complex
- P-CR
plant-conserved region
- CSR
class-specific region
- CatD
catalytic domain
- PDB
Protein Data Bank
- SAXS
small-angle x-ray scattering
- SEC
size-exclusion chromatography
- TEV
tobacco etch virus
- CD
circular dichroism
- Rg
radius of gyration
- SeMet
selenomethionine
- SAD
single-wavelength anomalous dispersion
Footnotes
This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (grant no. DE–SC0000997); this work used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science user facility operated for the Office of Science by Argonne National Laboratory (contract no. DE–AC02–06CH11357); crystallographic data were collected at GM/CA@APS, supported by the National Cancer Institute (grant no. ACB–12002) and the National Institute of General Medical Sciences (grant no. AGM–12006); the Macromolecular Crystallography Facility was supported by the Purdue Center for Cancer Research (core grant no. P30 CA023168); SAXS data collection utilized the BioCAT resource supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant no. 9 P41 GM103622).
Articles can be viewed without a subscription.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Chacón P, Merelo JJ, Morán F (1993) Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng 6: 383–390 [DOI] [PubMed] [Google Scholar]
- Appenzeller L, Doblin M, Barreiro R, Wang HY, Niu XM, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11: 287–299 [Google Scholar]
- Aslanidis C, de Jong PJ (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18: 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov II, Pittman JK, Turner SR (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284: 3833–3841 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. (2014) Structure and function of voltage-gated sodium channels at atomic resolution. Exp Physiol 99: 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, Dodson E (2002) Jolly SAD. Acta Crystallogr D Biol Crystallogr 58: 494–506 [DOI] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, Valkov E, Alon A, Fass D, Axelrod HL, et al. (2011) Improved molecular replacement by density- and energy-guided protein structure optimization. Nature 473: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti R, Stepanov S, Rosenbaum G, Barrea R, Black E, Gore D, Heurich R, Kondrashkina E, Kropf AJ, Wang S, et al. (2004) The BioCAT undulator beamline 18ID: a facility for biological non-crystalline diffraction and x-ray absorption spectroscopy at the Advanced Photon Source. J Synchrotron Radiat 11: 399–405 [DOI] [PubMed] [Google Scholar]
- Giddings TH Jr, Brower DL, Staehelin LA (1980) Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol 84: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Guillot A, Vernhettes S, Höfte H (2014) Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol 166: 1709–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE (2008) Structural specificity in coiled-coil interactions. Curr Opin Struct Biol 18: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JL Jr, Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr 39: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Nojiri M, Hishikawa Y, Togawa E, Romanovicz D, Brown RM Jr (2002) Biodirected epitaxial nanodeposition of polymers on oriented macromolecular templates. Proc Natl Acad Sci USA 99: 14008–14013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI (2001) Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34: 33–41 [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La D, Esquivel-Rodríguez J, Venkatraman V, Li B, Sael L, Ueng S, Ahrendt S, Kihara D (2009) 3D-SURFER: software for high-throughput protein surface comparison and analysis. Bioinformatics 25: 2843–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Li S, Du J, Bashline L, Gu Y (2013) Cellulose synthase INTERACTIVE3 regulates cellulose biosynthesis in both a microtubule-dependent and microtubule-independent manner in Arabidopsis. Plant Cell 25: 4912–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y (2012) Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31: 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoori N, Timmers J, Desprez T, Alvim-Kamei CL, Dees DCT, Vincken JP, Visser RGF, Höfte H, Vernhettes S, Trindade LM (2014) KORRIGAN1 interacts specifically with integral components of the cellulose synthase machinery. PLoS ONE 9: e112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43: D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Fischer M, Rich J, Chen HM, Withers SG, Zimmer J (2016) Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Brown RM Jr (1980) Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol 84: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540 [DOI] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O’Neill H, Roberts EM, et al. (2016) Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthase complex. Sci Rep 6: 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles DR, Romanovicz DK, Brown RM Jr (2001) Cellulose in cyanobacteria: origin of vascular plant cellulose synthase? Plant Physiol 127: 529–542 [PMC free article] [PubMed] [Google Scholar]
- Olek AT, Rayon C, Makowski L, Kim HR, Ciesielski P, Badger J, Paul LN, Ghosh S, Kihara D, Crowley M, et al. (2014) The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell 26: 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning BW, Hunter CT III, Tayengwa R, Eveland AL, Dugard CK, Olek AT, Vermerris W, Koch KE, McCarty DR, Davis MF, et al. (2009) Genetic resources for maize cell wall biology. Plant Physiol 151: 1703–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Biou V (1997) Treatment of multiwavelength anomalous diffraction data as a special case of multiple isomorphous replacement. Methods Enzymol 276: 538–557 [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl CA, Strauss CE, Misura KM, Baker D (2004) Protein structure prediction using Rosetta. Methods Enzymol 383: 66–93 [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW (1974) Chemical and biological evolution of nucleotide-binding protein. Nature 250: 194–199 [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM Jr, Fevre M, Geremia RA, Henrissat B (1995) Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol 177: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Lin FC, Brown RM Jr (1990) Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol 15: 673–683 [DOI] [PubMed] [Google Scholar]
- Sethaphong L, Davis JK, Slabaugh E, Singh A, Haigler CH, Yingling YG (2016) Prediction of the structures of the plant-specific regions of vascular plant cellulose synthases and correlated functional analysis. Cellulose 23: 145–161 [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I, Lewis TE, Cuff A, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees JG, et al. (2015) CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res 43: D376–D381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh E, Davis JK, Haigler CH, Yingling YG, Zimmer J (2014) Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci 19: 99–106 [DOI] [PubMed] [Google Scholar]
- Slabaugh E, Scavuzzo-Duggan T, Chaves A, Wilson L, Wilson C, Davis JK, Cosgrove DJ, Anderson CT, Roberts AW, Haigler CH (2016) The valine and lysine residues in the conserved FxVTxK motif are important for the function of phylogenetically distant plant cellulose synthases. Glycobiology 26: 509–519 [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MH (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Svergun DI. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG. (2008) Cellulose biosynthesis and deposition in higher plants. New Phytol 178: 239–252 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Gardiner JC, Whiteman R, Turner SR (2004) Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11: 329 [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, DiMaio F, Read RJ, Baker D, Bunkóczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N (2012) phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics 13: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RGF, Trindade LM (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583: 978–982 [DOI] [PubMed] [Google Scholar]
- Tung CH, Huang JW, Yang JM (2007) Kappa-alpha plot derived structural alphabet and BLOSUM-like substitution matrix for rapid search of protein structure database. Genome Biol 8: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, et al. (2016) A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers. Plant Physiol 170: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara CE, Carpita NC (2001) β-D-Glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1→3),(1→4)β-D-glucan synthase. Plant Mol Biol 47: 145–160 [PubMed] [Google Scholar]
- Whitmore L, Wallace BA (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32: W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW, et al. (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA 87: 8130–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH III, Freshour GD, Hahn MG, Ye ZH (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.